+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3pms | ||||||
|---|---|---|---|---|---|---|---|
| Title | Recombinant peptide: N-glycanase F (PNGase F) | ||||||
 Components Components | Peptide:N-glycosidase F | ||||||
 Keywords Keywords | HYDROLASE / Jelly Roll Fold / N-Glycanase | ||||||
| Function / homology |  Function and homology information Function and homology informationoxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen / peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase / peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity / hydrolase activity, acting on glycosyl bonds Similarity search - Function | ||||||
| Biological species |  Elizabethkingia meningoseptica (bacteria) Elizabethkingia meningoseptica (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.57 Å molecular replacement / Resolution: 1.57 Å | ||||||
 Authors Authors | Filitcheva, J. / Anderson, B.F. / Norris, G.E. | ||||||
 Citation Citation |  Journal: To be Published Journal: To be PublishedTitle: Recombinant peptide:N-glycanase F (PNGase F) Authors: Filitcheva, J. / Anderson, B.F. / Norris, G.E. | ||||||
| History |
|
- Structure visualization
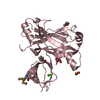
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3pms.cif.gz 3pms.cif.gz | 90 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3pms.ent.gz pdb3pms.ent.gz | 66.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3pms.json.gz 3pms.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  3pms_validation.pdf.gz 3pms_validation.pdf.gz | 467.4 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  3pms_full_validation.pdf.gz 3pms_full_validation.pdf.gz | 472.9 KB | Display | |
| Data in XML |  3pms_validation.xml.gz 3pms_validation.xml.gz | 19.5 KB | Display | |
| Data in CIF |  3pms_validation.cif.gz 3pms_validation.cif.gz | 29.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pm/3pms https://data.pdbj.org/pub/pdb/validation_reports/pm/3pms ftp://data.pdbj.org/pub/pdb/validation_reports/pm/3pms ftp://data.pdbj.org/pub/pdb/validation_reports/pm/3pms | HTTPS FTP |
-Related structure data
| Related structure data |  1pgsS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 36292.477 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Elizabethkingia meningoseptica (bacteria) Elizabethkingia meningoseptica (bacteria)Strain: CDC 3552 / Gene: png / Plasmid: OPH6 / Production host:  References: UniProt: Q9XBM8, peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| #2: Chemical | | #3: Chemical | ChemComp-GOL / #4: Chemical | #5: Water | ChemComp-HOH / | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.31 Å3/Da / Density % sol: 46.7 % |
|---|---|
| Crystal grow | Temperature: 295.15 K / Method: vapor diffusion, hanging drop / pH: 4.5 Details: 25% PEG 4000, 0.2M ammonium sulfate, 0.1M sodium acetate, pH 4.5, VAPOR DIFFUSION, HANGING DROP, temperature 295.15K |
-Data collection
| Diffraction | Mean temperature: 120 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU MICROMAX-007 / Wavelength: 1.542 Å ROTATING ANODE / Type: RIGAKU MICROMAX-007 / Wavelength: 1.542 Å |
| Detector | Type: RIGAKU RAXIS IV++ / Detector: IMAGE PLATE / Date: Jul 10, 2009 |
| Radiation | Monochromator: Capillary optics / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.542 Å / Relative weight: 1 |
| Reflection | Resolution: 1.57→40.41 Å / Num. all: 43449 / Num. obs: 41972 / % possible obs: 96.6 % / Rmerge(I) obs: 0.057 |
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1PGS Resolution: 1.57→34.02 Å / Cor.coef. Fo:Fc: 0.97 / Cor.coef. Fo:Fc free: 0.958 / Occupancy max: 1 / Occupancy min: 0.2 / SU B: 1.281 / SU ML: 0.047 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R Free: 0.076 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES: REFINED INDIVIDUALLY
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 55.05 Å2 / Biso mean: 18.3064 Å2 / Biso min: 6.03 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.57→34.02 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.57→1.611 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller














 PDBj
PDBj






