[English] 日本語
 Yorodumi
Yorodumi- PDB-3nyp: A bimolecular anti-parallel-stranded Oxytricha nova telomeric qua... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3nyp | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
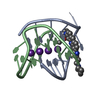
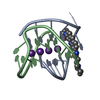
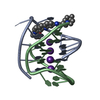
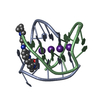
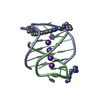
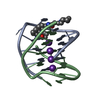
| Title | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine ligand containing bis-3-fluoropyrrolidine end side chains | ||||||||||||||||||
 Components Components | 5'-D(* Keywords KeywordsDNA / QUADRUPLEX / OXTYRICHA NOVA / BSU-6039 / BSU6039 / ANTI-PARALLEL / BIMOLECULAR / MACROMOLECULE / G-QUADRUPLEX / TELOMERE / ACRIDINE | Function / homology | : / Chem-RNR / DNA / DNA (> 10) |  Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.179 Å MOLECULAR REPLACEMENT / Resolution: 1.179 Å  Authors AuthorsCampbell, N.H. / Neidle, S. |  Citation Citation Journal: Org.Biomol.Chem. / Year: 2011 Journal: Org.Biomol.Chem. / Year: 2011Title: Fluorine in medicinal chemistry: beta-fluorination of peripheral pyrrolidines attached to acridine ligands affects their interactions with G-quadruplex DNA. Authors: Campbell, N.H. / Smith, D.L. / Reszka, A.P. / Neidle, S. / O'Hagan, D. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3nyp.cif.gz 3nyp.cif.gz | 44.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3nyp.ent.gz pdb3nyp.ent.gz | 32 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3nyp.json.gz 3nyp.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  3nyp_validation.pdf.gz 3nyp_validation.pdf.gz | 586.2 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  3nyp_full_validation.pdf.gz 3nyp_full_validation.pdf.gz | 592.8 KB | Display | |
| Data in XML |  3nyp_validation.xml.gz 3nyp_validation.xml.gz | 6.7 KB | Display | |
| Data in CIF |  3nyp_validation.cif.gz 3nyp_validation.cif.gz | 9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ny/3nyp https://data.pdbj.org/pub/pdb/validation_reports/ny/3nyp ftp://data.pdbj.org/pub/pdb/validation_reports/ny/3nyp ftp://data.pdbj.org/pub/pdb/validation_reports/ny/3nyp | HTTPS FTP |
-Related structure data
| Related structure data |  3nz7C  1l1hS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: DNA chain | Mass: 3805.460 Da / Num. of mol.: 2 / Source method: obtained synthetically Details: This telomeric sequence occurs naturally in the organism Oxytricha nova #2: Chemical | ChemComp-K / #3: Chemical | ChemComp-RNR / | #4: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.38 Å3/Da / Density % sol: 48.22 % |
|---|---|
| Crystal grow | Temperature: 285.15 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: The 5 Ul drop contained 1.2 mM quadruplex DNA, 1.2 mM ligand, 4% MPD, 4 mM potassium chloride, 4 mM magnesium chloride, 4 mM sodium chloride, 4 mM lithium sulphate, 3.2 mM spermine.4HCl and ...Details: The 5 Ul drop contained 1.2 mM quadruplex DNA, 1.2 mM ligand, 4% MPD, 4 mM potassium chloride, 4 mM magnesium chloride, 4 mM sodium chloride, 4 mM lithium sulphate, 3.2 mM spermine.4HCl and 16 mM potassium cacodylate buffer at pH 6.5. This was equilibrated against a reservoir well solution of 500 Ul of 45% MPD, VAPOR DIFFUSION, HANGING DROP, temperature 285.15K |
-Data collection
| Diffraction | Mean temperature: 105 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I04 / Wavelength: 0.9702 Å / Beamline: I04 / Wavelength: 0.9702 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Aug 9, 2009 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.9702 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.18→21.99 Å / Num. all: 23982 / Num. obs: 23982 / % possible obs: 97.7 % / Observed criterion σ(F): 2 / Observed criterion σ(I): 2 / Redundancy: 3.6 % / Rmerge(I) obs: 0.051 / Rsym value: 0.051 / Net I/σ(I): 15.5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 1L1H Resolution: 1.179→21.99 Å / Occupancy max: 1 / Occupancy min: 0.42 / SU ML: 0.13 / σ(F): 0.04 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 46.946 Å2 / ksol: 0.325 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 56.09 Å2 / Biso mean: 14.8626 Å2 / Biso min: 4.94 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.179→21.99 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 8
|
 Movie
Movie Controller
Controller










 PDBj
PDBj











































