[English] 日本語
 Yorodumi
Yorodumi- PDB-3eui: A bimolecular anti-parallel-stranded Oxytricha nova telomeric qua... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3eui | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6042 in a large unit cell | ||||||||||||||||||
 Components Components | 5'-D(* Keywords KeywordsDNA / quadruplex / Oxytricha nova / BSU-6042 / BSU6042 / anti-parallel / bimolecular / macromolecule / G-quadruplex | Function / homology | : / Chem-NC5 / Chem-NCI / SPERMINE / DNA / DNA (> 10) |  Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å MOLECULAR REPLACEMENT / Resolution: 2.2 Å  Authors AuthorsCampbell, N.H. / Parkinson, G. / Neidle, S. |  Citation Citation Journal: Biochemistry / Year: 2009 Journal: Biochemistry / Year: 2009Title: Selectivity in Ligand Recognition of G-Quadruplex Loops. Authors: Campbell, N.H. / Patel, M. / Tofa, A.B. / Ghosh, R. / Parkinson, G.N. / Neidle, S. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3eui.cif.gz 3eui.cif.gz | 46.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3eui.ent.gz pdb3eui.ent.gz | 32.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3eui.json.gz 3eui.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/eu/3eui https://data.pdbj.org/pub/pdb/validation_reports/eu/3eui ftp://data.pdbj.org/pub/pdb/validation_reports/eu/3eui ftp://data.pdbj.org/pub/pdb/validation_reports/eu/3eui | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3em2C  3eqwC  3eruC  3es0C 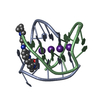 3et8C  3eumC  1l1hS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | The asymmetric unit contains two biological units. |
- Components
Components
-DNA chain , 1 types, 4 molecules ABCD
| #1: DNA chain | Mass: 3805.460 Da / Num. of mol.: 4 / Source method: obtained synthetically / Details: This sequence occurs naturally in Oxytricha nova |
|---|
-Non-polymers , 5 types, 173 molecules 








| #2: Chemical | ChemComp-K / #3: Chemical | ChemComp-NC5 / | #4: Chemical | ChemComp-SPM / | #5: Chemical | ChemComp-NCI / | #6: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.88 Å3/Da / Density % sol: 34.61 % | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 285.15 K / Method: vapor diffusion, hanging drop / pH: 7 Details: 2 microliter drops containing 5% v/v MPD, 0.50 mM DNA, 0.25 mM Ligand, 40 mM Potassium chloride, 5 mM Magnesium chloride, 4.1 Spermine equilibrated against 35% v/v MPD, pH 7.0, VAPOR ...Details: 2 microliter drops containing 5% v/v MPD, 0.50 mM DNA, 0.25 mM Ligand, 40 mM Potassium chloride, 5 mM Magnesium chloride, 4.1 Spermine equilibrated against 35% v/v MPD, pH 7.0, VAPOR DIFFUSION, HANGING DROP, temperature 285.15K | ||||||||||||||||||||||||
| Components of the solutions |
|
-Data collection
| Diffraction | Mean temperature: 105 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å |
| Detector | Type: RIGAKU RAXIS IV / Detector: IMAGE PLATE / Date: Jun 20, 2006 / Details: mirrors |
| Radiation | Monochromator: Osmic mirrors / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.2→27.72 Å / Num. all: 6193 / Num. obs: 5854 / % possible obs: 94.5 % / Observed criterion σ(I): 3 / Redundancy: 3.61 % / Biso Wilson estimate: 34.254 Å2 / Rmerge(I) obs: 0.087 / Net I/σ(I): 8.3 |
| Reflection shell | Resolution: 2.2→2.28 Å / Redundancy: 3.59 % / Rmerge(I) obs: 0.181 / Mean I/σ(I) obs: 5.2 / Num. unique all: 545 / % possible all: 92.1 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 1L1H Resolution: 2.2→27.68 Å / Cor.coef. Fo:Fc: 0.946 / Cor.coef. Fo:Fc free: 0.93 / SU B: 8.406 / SU ML: 0.225 / Cross valid method: THROUGHOUT / ESU R: 0.878 / ESU R Free: 0.311 / Stereochemistry target values: MAXIMUM LIKELIHOOD
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 20.772 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.2→27.68 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.2→2.257 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller










 PDBj
PDBj











































