[English] 日本語
 Yorodumi
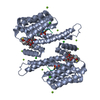
Yorodumi- PDB-3iqv: Crystal Structure of human 14-3-3 sigma in Complex with Raf1 pept... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3iqv | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of human 14-3-3 sigma in Complex with Raf1 peptide (6mer) and stabilisator Fusicoccin | ||||||
 Components Components |
| ||||||
 Keywords Keywords | PROTEIN BINDING / SIGNALING PROTEIN / SIGNAL TRANSDUCTION / Nucleus / Phosphoprotein / Secreted | ||||||
| Function / homology |  Function and homology information Function and homology informationdeath-inducing signaling complex assembly / intermediate filament cytoskeleton organization / regulation of Rho protein signal transduction / type B pancreatic cell proliferation / SHOC2 M1731 mutant abolishes MRAS complex function / Gain-of-function MRAS complexes activate RAF signaling / Rap1 signalling / insulin secretion involved in cellular response to glucose stimulus / Negative feedback regulation of MAPK pathway / IFNG signaling activates MAPKs ...death-inducing signaling complex assembly / intermediate filament cytoskeleton organization / regulation of Rho protein signal transduction / type B pancreatic cell proliferation / SHOC2 M1731 mutant abolishes MRAS complex function / Gain-of-function MRAS complexes activate RAF signaling / Rap1 signalling / insulin secretion involved in cellular response to glucose stimulus / Negative feedback regulation of MAPK pathway / IFNG signaling activates MAPKs / GP1b-IX-V activation signalling / regulation of epidermal cell division / protein kinase C inhibitor activity / positive regulation of epidermal cell differentiation / keratinocyte development / ERBB2-ERBB3 signaling pathway / keratinization / neurotrophin TRK receptor signaling pathway / face development / regulation of cell-cell adhesion / pseudopodium / regulation of cell differentiation / thyroid gland development / establishment of skin barrier / Regulation of localization of FOXO transcription factors / keratinocyte proliferation / extrinsic apoptotic signaling pathway via death domain receptors / somatic stem cell population maintenance / positive regulation of peptidyl-serine phosphorylation / Activation of BAD and translocation to mitochondria / phosphoserine residue binding / MAP kinase kinase kinase activity / negative regulation of keratinocyte proliferation / type II interferon-mediated signaling pathway / cAMP/PKA signal transduction / negative regulation of protein localization to plasma membrane / Schwann cell development / SARS-CoV-2 targets host intracellular signalling and regulatory pathways / negative regulation of protein kinase activity / negative regulation of extrinsic apoptotic signaling pathway via death domain receptors / negative regulation of stem cell proliferation / negative regulation of protein-containing complex assembly / SARS-CoV-1 targets host intracellular signalling and regulatory pathways / RHO GTPases activate PKNs / Chk1/Chk2(Cds1) mediated inactivation of Cyclin B:Cdk1 complex / positive regulation of protein localization / response to muscle stretch / myelination / CD209 (DC-SIGN) signaling / positive regulation of cell adhesion / insulin-like growth factor receptor signaling pathway / protein sequestering activity / negative regulation of innate immune response / TP53 Regulates Transcription of Genes Involved in G2 Cell Cycle Arrest / protein export from nucleus / release of cytochrome c from mitochondria / thymus development / positive regulation of protein export from nucleus / stem cell proliferation / adenylate cyclase activator activity / TP53 Regulates Metabolic Genes / Translocation of SLC2A4 (GLUT4) to the plasma membrane / wound healing / RAF activation / Signaling by high-kinase activity BRAF mutants / MAP2K and MAPK activation / Stimuli-sensing channels / intrinsic apoptotic signaling pathway in response to DNA damage / Negative regulation of MAPK pathway / Signaling by RAF1 mutants / Signaling by moderate kinase activity BRAF mutants / Paradoxical activation of RAF signaling by kinase inactive BRAF / Signaling downstream of RAS mutants / Signaling by BRAF and RAF1 fusions / insulin receptor signaling pathway / intracellular protein localization / sperm midpiece / MAPK cascade / regulation of protein localization / positive regulation of cell growth / regulation of apoptotic process / mitochondrial outer membrane / protein phosphorylation / protein kinase activity / non-specific serine/threonine protein kinase / positive regulation of MAPK cascade / regulation of cell cycle / cadherin binding / negative regulation of cell population proliferation / protein serine kinase activity / protein serine/threonine kinase activity / apoptotic process / protein kinase binding / negative regulation of apoptotic process / enzyme binding / negative regulation of transcription by RNA polymerase II / Golgi apparatus / signal transduction / positive regulation of transcription by RNA polymerase II / mitochondrion Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.2 Å MOLECULAR REPLACEMENT / Resolution: 1.2 Å | ||||||
 Authors Authors | Ottmann, C. / Weyand, M. | ||||||
 Citation Citation |  Journal: Mol.Cell.Biol. / Year: 2010 Journal: Mol.Cell.Biol. / Year: 2010Title: Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling. Authors: Molzan, M. / Schumacher, B. / Ottmann, C. / Baljuls, A. / Polzien, L. / Weyand, M. / Thiel, P. / Rose, R. / Rose, M. / Kuhenne, P. / Kaiser, M. / Rapp, U.R. / Kuhlmann, J. / Ottmann, C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3iqv.cif.gz 3iqv.cif.gz | 140.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3iqv.ent.gz pdb3iqv.ent.gz | 108.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3iqv.json.gz 3iqv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/iq/3iqv https://data.pdbj.org/pub/pdb/validation_reports/iq/3iqv ftp://data.pdbj.org/pub/pdb/validation_reports/iq/3iqv ftp://data.pdbj.org/pub/pdb/validation_reports/iq/3iqv | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3cu8C  3iqjC  3iquC  3nkxC  3o8iC  1ywtS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein / Protein/peptide , 2 types, 2 molecules AP
| #1: Protein | Mass: 26558.914 Da / Num. of mol.: 1 / Fragment: UNP residues 1-231 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: P31947 Homo sapiens (human) / References: UniProt: P31947 |
|---|---|
| #2: Protein/peptide | Mass: 759.680 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: The peptide was chemically synthesized. The sequence of the peptide is naturally found in Homo sapiens (human) References: UniProt: P04049 |
-Non-polymers , 4 types, 442 molecules 






| #3: Chemical | ChemComp-FSC / | ||||
|---|---|---|---|---|---|
| #4: Chemical | | #5: Chemical | ChemComp-MG / #6: Water | ChemComp-HOH / | |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.63 Å3/Da / Density % sol: 53.22 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 7.5 Details: 20mM Hepes/NaOH pH 7.5, 2mM MgCl2 and 2 mM DTT, set up for crystallization in 0.1 M Hepes/NaOH ph 7.5, 0.2 M CaCl2, 28% PEG 400, 5% glycerol, 2mM DTT, VAPOR DIFFUSION, HANGING DROP, temperature 277K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X10SA / Beamline: X10SA |
| Detector | Type: MARMOSAIC 225 mm CCD / Detector: CCD / Date: Feb 16, 2009 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Relative weight: 1 |
| Reflection | Resolution: 1.2→50 Å / Num. all: 90050 / Num. obs: 89939 / % possible obs: 99.9 % / Observed criterion σ(F): -3 / Observed criterion σ(I): -3 / Redundancy: 8.3 % / Biso Wilson estimate: 15.46 Å2 / Rsym value: 0.065 / Net I/σ(I): 19.1 |
| Reflection shell | Resolution: 1.2→1.25 Å / Redundancy: 7.9 % / Mean I/σ(I) obs: 4.4 / Num. unique all: 10234 / Rsym value: 0.551 / % possible all: 99.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1YWT Resolution: 1.2→41.65 Å / Cor.coef. Fo:Fc: 0.976 / Cor.coef. Fo:Fc free: 0.969 / SU B: 0.789 / SU ML: 0.017 / Cross valid method: THROUGHOUT / ESU R: 0.03 / ESU R Free: 0.032 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 10.554 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.2→41.65 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.2→1.231 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller










 PDBj
PDBj

















