+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3g9p | ||||||
|---|---|---|---|---|---|---|---|
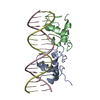
| Title | GR DNA binding domain:Sgk 16bp complex-7 | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSCRIPTION/DNA / glucocorticoid / DNA-binding / allostry / lever arm / hormone / Alternative initiation / Chromatin regulator / Cytoplasm / Lipid-binding / Metal-binding / Nucleus / Phosphoprotein / Polymorphism / Receptor / Steroid-binding / Transcription / Transcription regulation / Ubl conjugation / Zinc / Zinc-finger / TRANSCRIPTION-DNA COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationnuclear receptor-mediated corticosteroid signaling pathway / negative regulation of behavioral fear response / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / muscle atrophy / negative regulation of synaptic plasticity / positive regulation of nuclear receptor-mediated glucocorticoid signaling pathway / nuclear receptor-mediated glucocorticoid signaling pathway / negative regulation of long-term synaptic depression / response to inactivity / regulation of glucocorticoid biosynthetic process ...nuclear receptor-mediated corticosteroid signaling pathway / negative regulation of behavioral fear response / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / muscle atrophy / negative regulation of synaptic plasticity / positive regulation of nuclear receptor-mediated glucocorticoid signaling pathway / nuclear receptor-mediated glucocorticoid signaling pathway / negative regulation of long-term synaptic depression / response to inactivity / regulation of glucocorticoid biosynthetic process / nuclear glucocorticoid receptor activity / positive regulation of cell growth involved in cardiac muscle cell development / SUMOylation of intracellular receptors / Nuclear Receptor transcription pathway / steroid hormone binding / response to mercury ion / glucocorticoid metabolic process / response to cortisol / neuroinflammatory response / Leydig cell differentiation / mammary gland duct morphogenesis / microglia differentiation / response to arsenic-containing substance / cellular response to magnesium ion / maternal behavior / astrocyte differentiation / negative regulation of vascular permeability / positive regulation of glutamate secretion / adrenal gland development / regulation of gluconeogenesis / cellular response to glucocorticoid stimulus / response to corticosterone / cellular response to steroid hormone stimulus / response to dexamethasone / positive regulation of dendritic spine development / motor behavior / hormone binding / androgen metabolic process / regulation of glucose metabolic process / associative learning / response to electrical stimulus / estrogen response element binding / cellular response to dexamethasone stimulus / nuclear receptor-mediated steroid hormone signaling pathway / cellular response to transforming growth factor beta stimulus / postsynaptic density, intracellular component / core promoter sequence-specific DNA binding / steroid binding / heat shock protein binding / Hsp70 protein binding / transcription initiation-coupled chromatin remodeling / lung development / TBP-class protein binding / positive regulation of cytokine production / response to activity / RNA polymerase II transcription regulatory region sequence-specific DNA binding / synaptic transmission, glutamatergic / promoter-specific chromatin binding / Hsp90 protein binding / female pregnancy / circadian rhythm / response to calcium ion / response to insulin / receptor tyrosine kinase binding / response to wounding / positive regulation of miRNA transcription / DNA-binding transcription repressor activity, RNA polymerase II-specific / spindle / nuclear receptor activity / sequence-specific double-stranded DNA binding / positive regulation of neuron apoptotic process / regulation of cell population proliferation / protein-containing complex assembly / DNA-binding transcription activator activity, RNA polymerase II-specific / double-stranded DNA binding / gene expression / sequence-specific DNA binding / dendritic spine / transcription coactivator activity / nuclear speck / RNA polymerase II cis-regulatory region sequence-specific DNA binding / chromatin remodeling / DNA-binding transcription factor activity / negative regulation of DNA-templated transcription / chromatin binding / centrosome / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / protein kinase binding / negative regulation of apoptotic process / chromatin / protein-containing complex binding / glutamatergic synapse / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / protein-containing complex / mitochondrion / DNA binding / zinc ion binding / identical protein binding Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.652 Å MOLECULAR REPLACEMENT / Resolution: 1.652 Å | ||||||
 Authors Authors | Pufall, M.A. / Yamamoto, K.R. / Meijsing, S.H. | ||||||
 Citation Citation |  Journal: Science / Year: 2009 Journal: Science / Year: 2009Title: DNA binding site sequence directs glucocorticoid receptor structure and activity. Authors: Meijsing, S.H. / Pufall, M.A. / So, A.Y. / Bates, D.L. / Chen, L. / Yamamoto, K.R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3g9p.cif.gz 3g9p.cif.gz | 114.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3g9p.ent.gz pdb3g9p.ent.gz | 85.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3g9p.json.gz 3g9p.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/g9/3g9p https://data.pdbj.org/pub/pdb/validation_reports/g9/3g9p ftp://data.pdbj.org/pub/pdb/validation_reports/g9/3g9p ftp://data.pdbj.org/pub/pdb/validation_reports/g9/3g9p | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3fylC  3g6pC  3g6qC  3g6rC  3g6tC  3g6uC  3g8uC  3g8xC  3g97C  3g99C  3g9iC  3g9jC  3g9mC  3g9oC  1gluS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 9962.758 Da / Num. of mol.: 2 / Fragment: UNP residues 440-525 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: DNA chain | | Mass: 4897.203 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: Glucocorticoid receptor binding site: Sgk 16 base top strand #3: DNA chain | | Mass: 4897.203 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: Glucocorticoid receptor binding site: Sgk 16 base bottom strand #4: Chemical | ChemComp-ZN / #5: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.08 Å3/Da / Density % sol: 60.01 % | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 6 Details: 50mM MES, pH 6.0, 20mM MgCl2, 15% isopropanol, VAPOR DIFFUSION, HANGING DROP, temperature 298K | ||||||||||||||||||||||||
| Components of the solutions |
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 8.3.1 / Wavelength: 1.115878 Å / Beamline: 8.3.1 / Wavelength: 1.115878 Å |
| Detector | Type: ADSC QUANTUM 210 / Detector: CCD / Date: May 12, 2007 |
| Radiation | Monochromator: Double flat crystal, Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.115878 Å / Relative weight: 1 |
| Reflection | Resolution: 1.59→50 Å / Num. obs: 42923 / % possible obs: 88.2 % / Observed criterion σ(I): 0 / Redundancy: 8.1 % / Biso Wilson estimate: 23.068 Å2 / Rmerge(I) obs: 0.077 / Rsym value: 0.078 / Net I/σ(I): 19.5 |
| Reflection shell | Resolution: 1.59→1.65 Å / Redundancy: 4.2 % / Rmerge(I) obs: 0.644 / Mean I/σ(I) obs: 1.7 / Num. unique all: 1664 / Rsym value: 0.53 / % possible all: 34.6 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1GLU Resolution: 1.652→34.354 Å / SU ML: 1.4 / σ(F): 0.12 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 56.535 Å2 / ksol: 0.36 e/Å3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 50.2 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.652→34.354 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj









































