[English] 日本語
 Yorodumi
Yorodumi- PDB-3f1o: Crystal structure of the high affinity heterodimer of HIF2 alpha ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3f1o | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains, with an internally-bound artificial ligand | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSCRIPTION / PAS domain / heterodimer / internal cavity / Activator / Angiogenesis / Congenital erythrocytosis / Developmental protein / Differentiation / Disease mutation / DNA-binding / Hydroxylation / Nucleus / Phosphoprotein / Transcription regulation / Ubl conjugation / Alternative splicing / Polymorphism | ||||||
| Function / homology |  Function and homology information Function and homology informationmyoblast fate commitment / nuclear aryl hydrocarbon receptor complex / positive regulation of hormone biosynthetic process / Aryl hydrocarbon receptor signalling / aryl hydrocarbon receptor complex / Cellular response to hypoxia / PTK6 Expression / Transcriptional regulation of pluripotent stem cells / regulation of protein neddylation / positive regulation of protein sumoylation ...myoblast fate commitment / nuclear aryl hydrocarbon receptor complex / positive regulation of hormone biosynthetic process / Aryl hydrocarbon receptor signalling / aryl hydrocarbon receptor complex / Cellular response to hypoxia / PTK6 Expression / Transcriptional regulation of pluripotent stem cells / regulation of protein neddylation / positive regulation of protein sumoylation / norepinephrine metabolic process / Xenobiotics / surfactant homeostasis / Phase I - Functionalization of compounds / epithelial cell maturation / positive regulation of vascular endothelial growth factor receptor signaling pathway / Regulation of gene expression by Hypoxia-inducible Factor / aryl hydrocarbon receptor binding / positive regulation of vascular endothelial growth factor production / blood vessel remodeling / Endogenous sterols / embryonic placenta development / cis-regulatory region sequence-specific DNA binding / positive regulation of endothelial cell proliferation / NPAS4 regulates expression of target genes / visual perception / lung development / regulation of heart rate / positive regulation of erythrocyte differentiation / Pexophagy / positive regulation of glycolytic process / erythrocyte differentiation / RNA polymerase II transcription regulatory region sequence-specific DNA binding / mitochondrion organization / PPARA activates gene expression / mRNA transcription by RNA polymerase II / Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha / multicellular organismal-level iron ion homeostasis / negative regulation of inflammatory response / RNA polymerase II transcription regulator complex / transcription coactivator binding / nuclear receptor activity / sequence-specific double-stranded DNA binding / positive regulation of cold-induced thermogenesis / Neddylation / response to oxidative stress / DNA-binding transcription activator activity, RNA polymerase II-specific / cellular response to oxidative stress / angiogenesis / transcription regulator complex / cellular response to hypoxia / sequence-specific DNA binding / RNA polymerase II-specific DNA-binding transcription factor binding / DNA-binding transcription factor activity, RNA polymerase II-specific / response to hypoxia / cell differentiation / nuclear speck / nuclear body / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / protein heterodimerization activity / regulation of transcription by RNA polymerase II / chromatin / signal transduction / protein homodimerization activity / positive regulation of transcription by RNA polymerase II / nucleoplasm / nucleus / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.598 Å MOLECULAR REPLACEMENT / Resolution: 1.598 Å | ||||||
 Authors Authors | Scheuermann, T.H. / Tomchick, D.R. / Machius, M. / Guo, Y. / Bruick, R.K. / Gardner, K.H. | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2009 Journal: Proc.Natl.Acad.Sci.USA / Year: 2009Title: Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor. Authors: Scheuermann, T.H. / Tomchick, D.R. / Machius, M. / Guo, Y. / Bruick, R.K. / Gardner, K.H. | ||||||
| History |
| ||||||
| Remark 650 | HELIX DETERMINATION METHOD: AUTHOR DETERMINED | ||||||
| Remark 700 | SHEET DETERMINATION METHOD: AUTHOR DETERMINED |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3f1o.cif.gz 3f1o.cif.gz | 155.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3f1o.ent.gz pdb3f1o.ent.gz | 125.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3f1o.json.gz 3f1o.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/f1/3f1o https://data.pdbj.org/pub/pdb/validation_reports/f1/3f1o ftp://data.pdbj.org/pub/pdb/validation_reports/f1/3f1o ftp://data.pdbj.org/pub/pdb/validation_reports/f1/3f1o | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3f1nC  3f1pC  2b02S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 13538.300 Da / Num. of mol.: 1 / Fragment: HIF2 alpha C-terminal PAS domain / Mutation: R247E Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: EPAS1, HIF2A, Hypoxia Inducible Factor 2 alpha, MOP2 / Production host: Homo sapiens (human) / Gene: EPAS1, HIF2A, Hypoxia Inducible Factor 2 alpha, MOP2 / Production host:  |
|---|---|
| #2: Protein | Mass: 14243.098 Da / Num. of mol.: 1 / Fragment: ARNT C-terminal PAS domain / Mutation: E362R Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: ARNT, Aryl Hydrocarbon Receptor Nuclear Translocator / Production host: Homo sapiens (human) / Gene: ARNT, Aryl Hydrocarbon Receptor Nuclear Translocator / Production host:  |
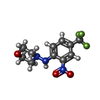
| #3: Chemical | ChemComp-2XY / |
| #4: Chemical | ChemComp-EDO / |
| #5: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.2 Å3/Da / Density % sol: 44.02 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion combined with microseeding / pH: 6.5 Details: 30% PEG 3350, 0.1M BisTris, 0.05M Tris, 0.017M NaCl, 0.005M DTT , pH 6.5, vapor diffusion combined with microseeding, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-BM / Wavelength: 0.97934 Å / Beamline: 19-BM / Wavelength: 0.97934 Å |
| Detector | Type: SBC-2 / Detector: CCD / Date: Jun 22, 2007 |
| Radiation | Monochromator: Custom / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97934 Å / Relative weight: 1 |
| Reflection | Resolution: 1.6→26.93 Å / Num. all: 31184 / Num. obs: 31184 / % possible obs: 98.4 % / Observed criterion σ(F): -3 / Observed criterion σ(I): -3 / Redundancy: 3.7 % / Biso Wilson estimate: 17.93 Å2 / Rmerge(I) obs: 0.05 / Net I/σ(I): 23.5 |
| Reflection shell | Resolution: 1.6→1.61 Å / Redundancy: 3.5 % / Rmerge(I) obs: 0.488 / Mean I/σ(I) obs: 2.1 / Num. unique all: 732 / % possible all: 97.7 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 2B02 Resolution: 1.598→26.93 Å / SU ML: 0.19 / Isotropic thermal model: isotropic / Cross valid method: THROUGHOUT / σ(F): 1.36 / Stereochemistry target values: ML Details: The density for the N463 side chain is relatively poor, that its conformation was informed by that observed in the higher resolution apo structure (rcsb050035, 3F1P) and modeled with a heavy ...Details: The density for the N463 side chain is relatively poor, that its conformation was informed by that observed in the higher resolution apo structure (rcsb050035, 3F1P) and modeled with a heavy weighting toward geometric ideality at the expense of a best fit against poor density
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 49.905 Å2 / ksol: 0.412 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 23.21 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.598→26.93 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller















 PDBj
PDBj