+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3enc | ||||||
|---|---|---|---|---|---|---|---|
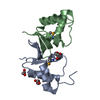
| Title | Crystal structure of Pyrococcus furiosus PCC1 dimer | ||||||
 Components Components | protein PCC1 | ||||||
 Keywords Keywords | UNKNOWN FUNCTION / dimerization domain / KEOPS / telomere | ||||||
| Function / homology | : / CTAG/Pcc1 family / Transcription factor Pcc1 / Alpha-D-phosphohexomutase, C-terminal domain / TATA-Binding Protein / 2-Layer Sandwich / Alpha Beta / KEOPS complex subunit Pcc1 Function and homology information Function and homology information | ||||||
| Biological species |   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SAD / Resolution: 2.6304 Å SAD / Resolution: 2.6304 Å | ||||||
 Authors Authors | Neculai, D. | ||||||
 Citation Citation |  Journal: Mol.Cell / Year: 2008 Journal: Mol.Cell / Year: 2008Title: Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine. Authors: Mao, D.Y. / Neculai, D. / Downey, M. / Orlicky, S. / Haffani, Y.Z. / Ceccarelli, D.F. / Ho, J.S. / Szilard, R.K. / Zhang, W. / Ho, C.S. / Wan, L. / Fares, C. / Rumpel, S. / Kurinov, I. / ...Authors: Mao, D.Y. / Neculai, D. / Downey, M. / Orlicky, S. / Haffani, Y.Z. / Ceccarelli, D.F. / Ho, J.S. / Szilard, R.K. / Zhang, W. / Ho, C.S. / Wan, L. / Fares, C. / Rumpel, S. / Kurinov, I. / Arrowsmith, C.H. / Durocher, D. / Sicheri, F. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3enc.cif.gz 3enc.cif.gz | 77.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3enc.ent.gz pdb3enc.ent.gz | 59.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3enc.json.gz 3enc.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/en/3enc https://data.pdbj.org/pub/pdb/validation_reports/en/3enc ftp://data.pdbj.org/pub/pdb/validation_reports/en/3enc ftp://data.pdbj.org/pub/pdb/validation_reports/en/3enc | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 10041.199 Da / Num. of mol.: 2 / Fragment: pcc1 / Mutation: I12M Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Pyrococcus furiosus (archaea) / Strain: DSM 3638 / Gene: PF2011 / Plasmid: pGEX / Production host: Pyrococcus furiosus (archaea) / Strain: DSM 3638 / Gene: PF2011 / Plasmid: pGEX / Production host:  Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.68 Å3/Da / Density % sol: 54.09 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 6.1 Details: 45% PEG 300 0.1 M Na/K phosphate, pH 6.1, VAPOR DIFFUSION, HANGING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  CHESS CHESS  / Beamline: F1 / Wavelength: 0.97935 Å / Beamline: F1 / Wavelength: 0.97935 Å |
| Detector | Type: ADSC QUANTUM 210 / Detector: CCD / Date: Nov 10, 2007 / Details: MIRRORS |
| Radiation | Monochromator: HORIZONTAL BENT SI(111), ASYMMETRICALLY CUT WITH WATER COOLED CU BLOCK Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97935 Å / Relative weight: 1 |
| Reflection twin | Type: pseudo-merohedral / Operator: h,-h-k,-l / Fraction: 0.503 |
| Reflection | Resolution: 2.6304→68.251 Å / Num. all: 6413 / Num. obs: 6401 / % possible obs: 99.8 % / Observed criterion σ(I): 2.48 / Redundancy: 1.93 % / Rmerge(I) obs: 0.0331 / Rsym value: 0.0305 / Net I/σ(I): 20.16 |
| Reflection shell | Resolution: 2.6304→2.72 Å / Redundancy: 1.93 % / Rmerge(I) obs: 0.2677 / Mean I/σ(I) obs: 2.86 / Num. unique all: 612 / Rsym value: 0.398 / % possible all: 100 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  SAD / Resolution: 2.6304→68.251 Å / Cross valid method: THROUGHOUT / σ(F): 2.06 / Stereochemistry target values: TWIN_LSQ_F SAD / Resolution: 2.6304→68.251 Å / Cross valid method: THROUGHOUT / σ(F): 2.06 / Stereochemistry target values: TWIN_LSQ_F
| ||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 138.453 Å2 / ksol: 0.356 e/Å3 | ||||||||||||||||||||||||||||||||||||||||
| Displacement parameters |
| ||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.6304→68.251 Å
| ||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: 14.1503 Å / Origin y: 24.3766 Å / Origin z: 30.0971 Å
| ||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group | Selection details: all |
 Movie
Movie Controller
Controller

















 PDBj
PDBj
