[English] 日本語
 Yorodumi
Yorodumi- PDB-3bmq: Structure of Pteridine Reductase 1 (PTR1) from Trypanosoma brucei... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3bmq | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of Pteridine Reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor (Compound AX5) | ||||||
 Components Components | (Pteridine reductase) x 2 | ||||||
 Keywords Keywords | OXIDOREDUCTASE / pteridine reductase / ptr1 / trypanosoma brucei / short chain dehydrogenase / inhibitor | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.7 Å molecular replacement / Resolution: 1.7 Å | ||||||
 Authors Authors | Martini, V.P. / Iulek, J. / Hunter, W.N. | ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2010 Journal: J.Med.Chem. / Year: 2010Title: Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases. Authors: Tulloch, L.B. / Martini, V.P. / Iulek, J. / Huggan, J.K. / Lee, J.H. / Gibson, C.L. / Smith, T.K. / Suckling, C.J. / Hunter, W.N. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3bmq.cif.gz 3bmq.cif.gz | 243.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3bmq.ent.gz pdb3bmq.ent.gz | 193.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3bmq.json.gz 3bmq.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bm/3bmq https://data.pdbj.org/pub/pdb/validation_reports/bm/3bmq ftp://data.pdbj.org/pub/pdb/validation_reports/bm/3bmq ftp://data.pdbj.org/pub/pdb/validation_reports/bm/3bmq | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3bmcC  3bmnC  3bmoC  3jq6C  3jq7C  3jq8C  3jq9C  3jqaC  3jqbC  3jqcC  3jqdC  3jqeC  3jqfC  3jqgC  3bms  3bmt C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 2 types, 4 molecules ADBC
| #1: Protein | Mass: 30685.787 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: Protein | Mass: 30669.791 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|
-Non-polymers , 6 types, 1376 molecules 
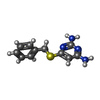









| #3: Chemical | ChemComp-NAP / #4: Chemical | ChemComp-AX5 / #5: Chemical | #6: Chemical | #7: Chemical | #8: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.07 Å3/Da / Density % sol: 40.62 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 4.5 Details: 2-3M sodium acetate, 10-100mM sodium citrate, pH 4.5, vapor diffusion, hanging drop, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SRS SRS  / Beamline: PX14.1 / Beamline: PX14.1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.7→74.625 Å / Num. obs: 94013 / % possible obs: 85.8 % / Redundancy: 5.6 % / Rmerge(I) obs: 0.055 / Rsym value: 0.055 / Net I/σ(I): 8.4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 1.7→74.54 Å / Cor.coef. Fo:Fc: 0.974 / Cor.coef. Fo:Fc free: 0.95 / SU B: 3.182 / SU ML: 0.059 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.102 / ESU R Free: 0.106 / Stereochemistry target values: MAXIMUM LIKELIHOOD MOLECULAR REPLACEMENT / Resolution: 1.7→74.54 Å / Cor.coef. Fo:Fc: 0.974 / Cor.coef. Fo:Fc free: 0.95 / SU B: 3.182 / SU ML: 0.059 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.102 / ESU R Free: 0.106 / Stereochemistry target values: MAXIMUM LIKELIHOODDetails: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS All of the 17 related structures are expressed from the same DNA construct, which encodes CYS at positions 59 and 168. Crystals harvested ...Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS All of the 17 related structures are expressed from the same DNA construct, which encodes CYS at positions 59 and 168. Crystals harvested within a couple of days of formation contain CYS at positions 59 and 168. However these two residues appear quite reactive and over time become oxidised to CSX, as determined by the emergence in older crystals of electron density for the OD atom. Sometimes CYS168 reacts with DTT in the crystallisation buffer, covalently linking the two molecules by an S-S bond.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 9.163 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.7→74.54 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.7→1.744 Å / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller






















 PDBj
PDBj












