| Entry | Database: PDB / ID: 2xz4
|
|---|
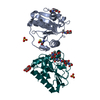
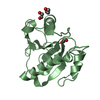
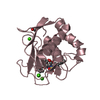
| Title | Crystal structure of the LFZ ectodomain of the peptidoglycan recognition protein LF |
|---|
 Components Components | PEPTIDOGLYCAN-RECOGNITION PROTEIN LF |
|---|
 Keywords Keywords | IMMUNE SYSTEM / INNATE IMMUNITY |
|---|
| Function / homology |  Function and homology information Function and homology information
Antimicrobial peptides / negative regulation of peptidoglycan recognition protein signaling pathway / defense response to other organism / negative regulation of immune response / response to bacterium / antibacterial humoral response / signaling receptor binding / innate immune response / zinc ion binding / plasma membraneSimilarity search - Function Peptidoglycan recognition protein family domain, metazoa/bacteria / Peptidoglycan recognition protein / Animal peptidoglycan recognition proteins homologous to Bacteriophage T3 lysozyme. / Lysozyme-like / Peptidoglycan recognition protein-like / N-acetylmuramoyl-L-alanine amidase / Ami_2 / N-acetylmuramoyl-L-alanine amidase domain / N-acetylmuramoyl-L-alanine amidase/PGRP domain superfamily / 3-Layer(aba) Sandwich / Alpha BetaSimilarity search - Domain/homology |
|---|
| Biological species |   DROSOPHILA MELANOGASTER (fruit fly) DROSOPHILA MELANOGASTER (fruit fly) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.72 Å MOLECULAR REPLACEMENT / Resolution: 1.72 Å |
|---|
 Authors Authors | Basbous, N. / Coste, F. / Leone, P. / Vincentelli, R. / Royet, J. / Kellenberger, C. / Roussel, A. |
|---|
 Citation Citation |  Journal: Embo Rep. / Year: 2011 Journal: Embo Rep. / Year: 2011
Title: The Drosophila Peptidoglycan-Recognition Protein Lf Interacts with Peptidoglycan-Recognition Protein Lc to Downregulate the Imd Pathway.
Authors: Basbous, N. / Coste, F. / Leone, P. / Vincentelli, R. / Royet, J. / Kellenberger, C. / Roussel, A. |
|---|
| History | | Deposition | Nov 23, 2010 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Apr 13, 2011 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | May 8, 2011 | Group: Version format compliance |
|---|
| Revision 1.2 | Jul 13, 2011 | Group: Version format compliance |
|---|
| Revision 1.3 | Dec 20, 2023 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Other / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_database_status / pdbx_initial_refinement_model / struct_conn / struct_site
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_database_status.status_code_sf / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr1_symmetry / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_conn.ptnr2_symmetry / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id |
|---|
| Revision 1.4 | Nov 13, 2024 | Group: Structure summary / Category: pdbx_entry_details / pdbx_modification_feature / Item: _pdbx_entry_details.has_protein_modification |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.72 Å
MOLECULAR REPLACEMENT / Resolution: 1.72 Å  Authors
Authors Citation
Citation Journal: Embo Rep. / Year: 2011
Journal: Embo Rep. / Year: 2011 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2xz4.cif.gz
2xz4.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2xz4.ent.gz
pdb2xz4.ent.gz PDB format
PDB format 2xz4.json.gz
2xz4.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/xz/2xz4
https://data.pdbj.org/pub/pdb/validation_reports/xz/2xz4 ftp://data.pdbj.org/pub/pdb/validation_reports/xz/2xz4
ftp://data.pdbj.org/pub/pdb/validation_reports/xz/2xz4
 Links
Links Assembly
Assembly

 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ESRF
ESRF  / Beamline: ID14-1 / Wavelength: 0.933
/ Beamline: ID14-1 / Wavelength: 0.933  Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller











 PDBj
PDBj