[English] 日本語
 Yorodumi
Yorodumi- PDB-2vif: Crystal structure of SOCS6 SH2 domain in complex with a c-KIT pho... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2vif | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of SOCS6 SH2 domain in complex with a c-KIT phosphopeptide | ||||||
 Components Components |
| ||||||
 Keywords Keywords | SIGNALING PROTEIN / GROWTH REGULATION / SIGNAL TRANSDUCTION INHIBITOR / KIT REGULATOR / PHOSPHOTYROSINE | ||||||
| Function / homology |  Function and homology information Function and homology informationDasatinib-resistant KIT mutants / Imatinib-resistant KIT mutants / KIT mutants bind TKIs / Masitinib-resistant KIT mutants / Nilotinib-resistant KIT mutants / Regorafenib-resistant KIT mutants / Signaling by kinase domain mutants of KIT / Sunitinib-resistant KIT mutants / Signaling by juxtamembrane domain KIT mutants / Sorafenib-resistant KIT mutants ...Dasatinib-resistant KIT mutants / Imatinib-resistant KIT mutants / KIT mutants bind TKIs / Masitinib-resistant KIT mutants / Nilotinib-resistant KIT mutants / Regorafenib-resistant KIT mutants / Signaling by kinase domain mutants of KIT / Sunitinib-resistant KIT mutants / Signaling by juxtamembrane domain KIT mutants / Sorafenib-resistant KIT mutants / Signaling by extracellular domain mutants of KIT / hematopoietic stem cell migration / melanocyte adhesion / positive regulation of pyloric antrum smooth muscle contraction / positive regulation of colon smooth muscle contraction / stem cell factor receptor activity / positive regulation of vascular associated smooth muscle cell differentiation / melanocyte migration / Kit signaling pathway / tongue development / : / positive regulation of small intestine smooth muscle contraction / positive regulation of dendritic cell cytokine production / mast cell chemotaxis / positive regulation of mast cell proliferation / mast cell differentiation / Fc receptor signaling pathway / glycosphingolipid metabolic process / mast cell proliferation / positive regulation of long-term neuronal synaptic plasticity / positive regulation of pseudopodium assembly / positive regulation of mast cell cytokine production / negative regulation of T cell activation / lymphoid progenitor cell differentiation / detection of mechanical stimulus involved in sensory perception of sound / melanocyte differentiation / immature B cell differentiation / germ cell migration / erythropoietin-mediated signaling pathway / myeloid progenitor cell differentiation / regulation of growth / digestive tract development / negative regulation of programmed cell death / embryonic hemopoiesis / lamellipodium assembly / positive regulation of tyrosine phosphorylation of STAT protein / Regulation of KIT signaling / megakaryocyte development / growth factor binding / mast cell degranulation / stem cell population maintenance / cytokine binding / positive regulation of Notch signaling pathway / pigmentation / negative regulation of reproductive process / negative regulation of developmental process / hemopoiesis / immunological synapse / T cell differentiation / somatic stem cell population maintenance / spermatid development / cell surface receptor signaling pathway via JAK-STAT / response to cadmium ion / negative regulation of signal transduction / ectopic germ cell programmed cell death / hematopoietic progenitor cell differentiation / : / ovarian follicle development / proteasomal protein catabolic process / TFAP2 (AP-2) family regulates transcription of growth factors and their receptors / transmembrane receptor protein tyrosine kinase activity / signaling adaptor activity / Transcriptional and post-translational regulation of MITF-M expression and activity / Negative regulation of FLT3 / acrosomal vesicle / SH2 domain binding / B cell differentiation / Signaling by phosphorylated juxtamembrane, extracellular and kinase domain KIT mutants / epithelial cell proliferation / erythrocyte differentiation / cell chemotaxis / positive regulation of receptor signaling pathway via JAK-STAT / stem cell differentiation / defense response / Signaling by SCF-KIT / receptor protein-tyrosine kinase / visual learning / cytoplasmic side of plasma membrane / male gonad development / fibrillar center / cytokine-mediated signaling pathway / Constitutive Signaling by Aberrant PI3K in Cancer / cell-cell junction / cell migration / PIP3 activates AKT signaling / regulation of cell shape / regulation of cell population proliferation / protein autophosphorylation / Neddylation / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling Similarity search - Function | ||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.45 Å MOLECULAR REPLACEMENT / Resolution: 1.45 Å | ||||||
 Authors Authors | Bullock, A. / Pike, A.C.W. / Savitsky, P. / Keates, T. / Pilka, E.S. / von Delft, F. / Edwards, A. / Weigelt, J. / Arrowsmith, C.H. / Knapp, S. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2011 Journal: J.Biol.Chem. / Year: 2011Title: Structural Basis for C-Kit Inhibition by the Suppressor of Cytokine Signaling 6 (Socs6) Ubiquitin Ligase. Authors: Zadjali, F. / Pike, A.C.W. / Vesterlund, M. / Sun, J. / Wu, C. / Li, S.S. / Ronnstrand, L. / Knapp, S. / Bullock, A. / Flores-Morales, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2vif.cif.gz 2vif.cif.gz | 78.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2vif.ent.gz pdb2vif.ent.gz | 58.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2vif.json.gz 2vif.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2vif_validation.pdf.gz 2vif_validation.pdf.gz | 446.1 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2vif_full_validation.pdf.gz 2vif_full_validation.pdf.gz | 446.7 KB | Display | |
| Data in XML |  2vif_validation.xml.gz 2vif_validation.xml.gz | 9.2 KB | Display | |
| Data in CIF |  2vif_validation.cif.gz 2vif_validation.cif.gz | 12.6 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vi/2vif https://data.pdbj.org/pub/pdb/validation_reports/vi/2vif ftp://data.pdbj.org/pub/pdb/validation_reports/vi/2vif ftp://data.pdbj.org/pub/pdb/validation_reports/vi/2vif | HTTPS FTP |
-Related structure data
| Related structure data |  2izvS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 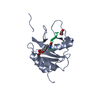
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 15918.718 Da / Num. of mol.: 1 / Fragment: SH2 DOMAIN, RESIDUES 361-499 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Plasmid: PNIC28-BSA4 / Production host: HOMO SAPIENS (human) / Plasmid: PNIC28-BSA4 / Production host:  |
|---|---|
| #2: Protein/peptide | Mass: 1349.297 Da / Num. of mol.: 1 / Fragment: RESIDUES 564-574 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Plasmid: PNIC28-BSA4 / Production host: HOMO SAPIENS (human) / Plasmid: PNIC28-BSA4 / Production host:  |
| #3: Chemical | ChemComp-EDO / |
| #4: Water | ChemComp-HOH / |
| Has protein modification | Y |
| Sequence details | TWO CLONING ARTIFACTS - V368D AND R463G |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.04 Å3/Da / Density % sol: 39.84 % / Description: NONE |
|---|---|
| Crystal grow | pH: 6.5 Details: 0.2M AMMONIUM SULPHATE, 30% PEG5000 MONOMETHYLETHER, 0.1M MES PH6.5 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X10SA / Wavelength: 0.98248 / Beamline: X10SA / Wavelength: 0.98248 |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Oct 26, 2007 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.98248 Å / Relative weight: 1 |
| Reflection | Resolution: 1.45→46.03 Å / Num. obs: 23956 / % possible obs: 99.9 % / Redundancy: 4.5 % / Biso Wilson estimate: 14.8 Å2 / Rmerge(I) obs: 0.089 / Net I/σ(I): 10.7 |
| Reflection shell | Resolution: 1.45→1.53 Å / Redundancy: 3.6 % / Rmerge(I) obs: 0.704 / Mean I/σ(I) obs: 3 / % possible all: 99.1 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2IZV Resolution: 1.45→35.69 Å / Cor.coef. Fo:Fc: 0.974 / Cor.coef. Fo:Fc free: 0.958 / SU B: 2.461 / SU ML: 0.044 / Cross valid method: THROUGHOUT / ESU R: 0.071 / ESU R Free: 0.07 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 14.05 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.45→35.69 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller


















 PDBj
PDBj














