[English] 日本語
 Yorodumi
Yorodumi- PDB-2v7o: Crystal structure of human calcium-calmodulin-dependent protein k... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2v7o | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of human calcium-calmodulin-dependent protein kinase II gamma | ||||||
 Components Components | CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE TYPE II GAMMA CHAIN | ||||||
 Keywords Keywords | TRANSFERASE / KINASE / S/T KINASE / ATP-BINDING / SERINE/THREONINE-PROTEIN KINASE / NUCLEOTIDE-BINDING / PHOSPHORYLATION / CALMODULIN-BINDING | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of skeletal muscle adaptation / calcium-dependent protein serine/threonine phosphatase activity / calcium- and calmodulin-dependent protein kinase complex / Ca2+/calmodulin-dependent protein kinase / Trafficking of AMPA receptors / calcium/calmodulin-dependent protein kinase activity / Assembly and cell surface presentation of NMDA receptors / insulin secretion / CREB1 phosphorylation through the activation of CaMKII/CaMKK/CaMKIV cascasde / CaMK IV-mediated phosphorylation of CREB ...regulation of skeletal muscle adaptation / calcium-dependent protein serine/threonine phosphatase activity / calcium- and calmodulin-dependent protein kinase complex / Ca2+/calmodulin-dependent protein kinase / Trafficking of AMPA receptors / calcium/calmodulin-dependent protein kinase activity / Assembly and cell surface presentation of NMDA receptors / insulin secretion / CREB1 phosphorylation through the activation of CaMKII/CaMKK/CaMKIV cascasde / CaMK IV-mediated phosphorylation of CREB / regulation of neuron projection development / Phase 0 - rapid depolarisation / Negative regulation of NMDA receptor-mediated neuronal transmission / Unblocking of NMDA receptors, glutamate binding and activation / regulation of calcium ion transport / Ion transport by P-type ATPases / Long-term potentiation / Regulation of MECP2 expression and activity / regulation of neuronal synaptic plasticity / HSF1-dependent transactivation / regulation of protein localization to plasma membrane / Ion homeostasis / sarcoplasmic reticulum membrane / Ras activation upon Ca2+ influx through NMDA receptor / RAF activation / long-term synaptic potentiation / Interferon gamma signaling / Signaling by RAF1 mutants / Signaling by moderate kinase activity BRAF mutants / Paradoxical activation of RAF signaling by kinase inactive BRAF / Signaling downstream of RAS mutants / endocytic vesicle membrane / Signaling by BRAF and RAF1 fusions / nervous system development / RAF/MAP kinase cascade / cell differentiation / calmodulin binding / neuron projection / postsynaptic density / protein serine kinase activity / protein homodimerization activity / nucleoplasm / ATP binding / identical protein binding / membrane / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.25 Å MOLECULAR REPLACEMENT / Resolution: 2.25 Å | ||||||
 Authors Authors | Pike, A.C.W. / Rellos, P. / Fedorov, O. / Burgess-Brown, N. / Shrestha, L. / Ugochukwu, E. / Pilka, E.S. / von Delft, F. / Edwards, A. / Weigelt, J. ...Pike, A.C.W. / Rellos, P. / Fedorov, O. / Burgess-Brown, N. / Shrestha, L. / Ugochukwu, E. / Pilka, E.S. / von Delft, F. / Edwards, A. / Weigelt, J. / Arrowsmith, C.H. / Sundstrom, M. / Knapp, S. | ||||||
 Citation Citation |  Journal: Plos Biol. / Year: 2010 Journal: Plos Biol. / Year: 2010Title: Structure of the Camkiidelta/Calmodulin Complex Reveals the Molecular Mechanism of Camkii Kinase Activation. Authors: Rellos, P. / Pike, A.C.W. / Niesen, F.H. / Salah, E. / Lee, W.H. / von Delft, F. / Knapp, S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2v7o.cif.gz 2v7o.cif.gz | 79.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2v7o.ent.gz pdb2v7o.ent.gz | 58 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2v7o.json.gz 2v7o.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2v7o_validation.pdf.gz 2v7o_validation.pdf.gz | 788.5 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2v7o_full_validation.pdf.gz 2v7o_full_validation.pdf.gz | 791.9 KB | Display | |
| Data in XML |  2v7o_validation.xml.gz 2v7o_validation.xml.gz | 14.9 KB | Display | |
| Data in CIF |  2v7o_validation.cif.gz 2v7o_validation.cif.gz | 20.5 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/v7/2v7o https://data.pdbj.org/pub/pdb/validation_reports/v7/2v7o ftp://data.pdbj.org/pub/pdb/validation_reports/v7/2v7o ftp://data.pdbj.org/pub/pdb/validation_reports/v7/2v7o | HTTPS FTP |
-Related structure data
| Related structure data |  2ux0C  2vn9C  2vz6C  2w2cC  2welC 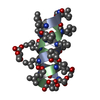 1bdwS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 38501.922 Da / Num. of mol.: 1 / Fragment: KINASE DOMAIN, RESIDUES 5-315 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Plasmid: PNIC28-BSA4 / Production host: HOMO SAPIENS (human) / Plasmid: PNIC28-BSA4 / Production host:  References: UniProt: Q8N4I3, UniProt: Q13555*PLUS, Ca2+/calmodulin-dependent protein kinase | ||||||
|---|---|---|---|---|---|---|---|
| #2: Chemical | ChemComp-DRN / | ||||||
| #3: Chemical | | #4: Water | ChemComp-HOH / | Has protein modification | Y | Nonpolymer details | BISINDOLYL | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.59 Å3/Da / Density % sol: 50.7 % / Description: NONE |
|---|---|
| Crystal grow | pH: 7 Details: 0.10M MAGNESIUM CHLORIDE, 0.1M HEPES PH7, 20% PEG6K, 10% ETHYLENE GLYCOL |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X10SA / Wavelength: 1.00721 / Beamline: X10SA / Wavelength: 1.00721 |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Jul 15, 2007 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.00721 Å / Relative weight: 1 |
| Reflection | Resolution: 2.25→42.33 Å / Num. obs: 18326 / % possible obs: 100 % / Observed criterion σ(I): 0 / Redundancy: 4 % / Rmerge(I) obs: 0.11 / Net I/σ(I): 10.4 |
| Reflection shell | Resolution: 2.25→2.37 Å / Redundancy: 4.1 % / Rmerge(I) obs: 0.72 / Mean I/σ(I) obs: 1.9 / % possible all: 100 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1BDW Resolution: 2.25→35.62 Å / Cor.coef. Fo:Fc: 0.96 / Cor.coef. Fo:Fc free: 0.927 / SU B: 14.48 / SU ML: 0.16 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / ESU R: 0.243 / ESU R Free: 0.213 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 33.45 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.25→35.62 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj