[English] 日本語
 Yorodumi
Yorodumi- PDB-2nvd: Human Aldose Reductase complexed with novel naphtho[1,2-d]isothia... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2nvd | ||||||
|---|---|---|---|---|---|---|---|
| Title | Human Aldose Reductase complexed with novel naphtho[1,2-d]isothiazole acetic acid derivative (2) | ||||||
 Components Components | Aldose reductase | ||||||
 Keywords Keywords | OXIDOREDUCTASE / aldose reductase / TIM-barrel / protein-ligand complex | ||||||
| Function / homology |  Function and homology information Function and homology informationglyceraldehyde oxidoreductase activity / Fructose biosynthesis / fructose biosynthetic process / L-glucuronate reductase activity / aldose reductase / D/L-glyceraldehyde reductase / glycerol dehydrogenase (NADP+) activity / C21-steroid hormone biosynthetic process / NADP-retinol dehydrogenase / Pregnenolone biosynthesis ...glyceraldehyde oxidoreductase activity / Fructose biosynthesis / fructose biosynthetic process / L-glucuronate reductase activity / aldose reductase / D/L-glyceraldehyde reductase / glycerol dehydrogenase (NADP+) activity / C21-steroid hormone biosynthetic process / NADP-retinol dehydrogenase / Pregnenolone biosynthesis / allyl-alcohol dehydrogenase / allyl-alcohol dehydrogenase activity / Galactose catabolism / prostaglandin H2 endoperoxidase reductase activity / regulation of urine volume / all-trans-retinol dehydrogenase (NADP+) activity / metanephric collecting duct development / daunorubicin metabolic process / doxorubicin metabolic process / retinal dehydrogenase (NAD+) activity / aldose reductase (NADPH) activity / epithelial cell maturation / cellular hyperosmotic salinity response / retinoid metabolic process / renal water homeostasis / carbohydrate metabolic process / electron transfer activity / negative regulation of apoptotic process / mitochondrion / extracellular space / extracellular exosome / nucleoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.55 Å MOLECULAR REPLACEMENT / Resolution: 1.55 Å | ||||||
 Authors Authors | Steuber, H. / Heine, A. / Klebe, G. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2007 Journal: J.Mol.Biol. / Year: 2007Title: Evidence for a novel binding site conformer of aldose reductase in ligand-bound state Authors: Steuber, H. / Zentgraf, M. / La Motta, C. / Sartini, S. / Heine, A. / Klebe, G. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2nvd.cif.gz 2nvd.cif.gz | 156.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2nvd.ent.gz pdb2nvd.ent.gz | 121.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2nvd.json.gz 2nvd.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/nv/2nvd https://data.pdbj.org/pub/pdb/validation_reports/nv/2nvd ftp://data.pdbj.org/pub/pdb/validation_reports/nv/2nvd ftp://data.pdbj.org/pub/pdb/validation_reports/nv/2nvd | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2nvcC  1el3S C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 35898.340 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: hALR2 / Plasmid: pET15b / Production host: Homo sapiens (human) / Gene: hALR2 / Plasmid: pET15b / Production host:  | ||
|---|---|---|---|
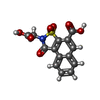
| #2: Chemical | ChemComp-NAP / | ||
| #3: Chemical | | #4: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.15 Å3/Da / Density % sol: 42.7 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 5 Details: 120mM ammonium citrate, 20% PEG 6000, pH 5.0, VAPOR DIFFUSION, HANGING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU300 / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU RU300 / Wavelength: 1.5418 Å |
| Detector | Type: RIGAKU RAXIS IV / Detector: IMAGE PLATE / Date: Jan 8, 2006 / Details: mirrors |
| Radiation | Monochromator: graphite / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.55→50 Å / Num. all: 42162 / Num. obs: 42162 / % possible obs: 94.9 % / Redundancy: 3.5 % / Rmerge(I) obs: 0.035 / Rsym value: 0.035 / Net I/σ(I): 39.3 |
| Reflection shell | Resolution: 1.55→1.58 Å / Redundancy: 2.3 % / Rmerge(I) obs: 0.169 / Mean I/σ(I) obs: 5.3 / Num. unique all: 1413 / Rsym value: 0.169 / % possible all: 63.7 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1EL3 Resolution: 1.55→30 Å / Num. parameters: 26284 / Num. restraintsaints: 31907 / Cross valid method: FREE R / σ(F): 0 / σ(I): 0 / Stereochemistry target values: ENGH & HUBER Details: ANISOTROPIC SCALING APPLIED BY THE METHOD OF PARKIN, MOEZZI & HOPE, J.APPL.CRYST.28(1995)53-56 ANISOTROPIC REFINEMENT REDUCED FREE R (NO CUTOFF) BY ?
| |||||||||||||||||||||||||||||||||
| Refine analyze | Num. disordered residues: 15 / Occupancy sum hydrogen: 2521 / Occupancy sum non hydrogen: 2850 | |||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.55→30 Å
| |||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj