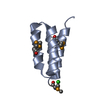
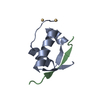
Entry Database : PDB / ID : 2n0zTitle Solution structure of MyUb (1080-1122) of human Myosin VI Unconventional myosin-VI Keywords / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / Model details lowest energy, model1 Authors He, F. / Walters, K. Funding support Organization Grant number Country National Institutes of Health/National Cancer Institute (NIH/NCI) CA136472 National Institutes of Health/National Cancer Institute (NIH/NCI) 1ZIABC011627
Journal : Cell Rep / Year : 2016Title : Myosin VI Contains a Compact Structural Motif that Binds to Ubiquitin Chains.Authors : He, F. / Wollscheid, H.P. / Nowicka, U. / Biancospino, M. / Valentini, E. / Ehlinger, A. / Acconcia, F. / Magistrati, E. / Polo, S. / Walters, K.J. History Deposition Mar 20, 2015 Deposition site / Processing site Revision 1.0 Mar 9, 2016 Provider / Type Revision 1.1 Mar 30, 2016 Group Revision 1.2 Apr 6, 2016 Group Revision 1.3 Apr 27, 2016 Group Revision 1.4 Oct 13, 2021 Group / Database referencesCategory database_2 / pdbx_nmr_software ... database_2 / pdbx_nmr_software / pdbx_nmr_spectrometer / struct_ref_seq_dif Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_nmr_software.name / _pdbx_nmr_spectrometer.model / _struct_ref_seq_dif.details Revision 2.0 Oct 20, 2021 Group Advisory / Atomic model ... Advisory / Atomic model / Author supporting evidence / Data collection / Database references / Derived calculations / Experimental preparation / Other / Refinement description / Source and taxonomy / Structure summary Category atom_site / entity_name_com ... atom_site / entity_name_com / entity_src_gen / pdbx_audit_support / pdbx_database_related / pdbx_database_status / pdbx_nmr_ensemble / pdbx_nmr_exptl / pdbx_nmr_exptl_sample / pdbx_nmr_exptl_sample_conditions / pdbx_nmr_refine / pdbx_nmr_sample_details / pdbx_nmr_software / pdbx_nmr_spectrometer / pdbx_poly_seq_scheme / pdbx_unobs_or_zero_occ_residues / pdbx_validate_close_contact / pdbx_validate_torsion / struct / struct_conf / struct_ref / struct_ref_seq / struct_ref_seq_dif Item _entity_src_gen.gene_src_common_name / _entity_src_gen.pdbx_beg_seq_num ... _entity_src_gen.gene_src_common_name / _entity_src_gen.pdbx_beg_seq_num / _entity_src_gen.pdbx_end_seq_num / _entity_src_gen.pdbx_gene_src_gene / _entity_src_gen.pdbx_seq_type / _pdbx_database_status.SG_entry / _pdbx_database_status.deposit_site / _pdbx_nmr_ensemble.conformer_selection_criteria / _pdbx_nmr_ensemble.conformers_calculated_total_number / _pdbx_nmr_exptl.sample_state / _pdbx_nmr_exptl.spectrometer_id / _pdbx_nmr_exptl_sample.component / _pdbx_nmr_exptl_sample_conditions.ionic_strength_units / _pdbx_nmr_exptl_sample_conditions.label / _pdbx_nmr_exptl_sample_conditions.pH_units / _pdbx_nmr_refine.software_ordinal / _pdbx_nmr_sample_details.contents / _pdbx_nmr_sample_details.label / _pdbx_nmr_sample_details.type / _pdbx_nmr_software.authors / _pdbx_nmr_software.classification / _pdbx_nmr_software.name / _pdbx_nmr_software.ordinal / _pdbx_nmr_software.version / _pdbx_nmr_spectrometer.type / _pdbx_poly_seq_scheme.auth_mon_id / _pdbx_poly_seq_scheme.auth_seq_num / _pdbx_poly_seq_scheme.pdb_mon_id / _struct.pdbx_CASP_flag / _struct_conf.beg_auth_comp_id / _struct_conf.beg_auth_seq_id / _struct_conf.beg_label_comp_id / _struct_conf.beg_label_seq_id / _struct_conf.end_auth_comp_id / _struct_conf.end_auth_seq_id / _struct_conf.end_label_comp_id / _struct_conf.end_label_seq_id / _struct_conf.pdbx_PDB_helix_id / _struct_conf.pdbx_PDB_helix_length / _struct_ref.db_code / _struct_ref.pdbx_align_begin / _struct_ref.pdbx_db_accession / _struct_ref_seq.db_align_beg / _struct_ref_seq.db_align_end / _struct_ref_seq.pdbx_db_accession / _struct_ref_seq_dif.pdbx_seq_db_accession_code Description / Provider / Type Revision 2.1 Jun 14, 2023 Group / Category / Item Revision 2.2 May 15, 2024 Group / Database references / Category / chem_comp_bond / database_2 / Item
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) Authors
Authors United States, 2items
United States, 2items  Citation
Citation Journal: Cell Rep / Year: 2016
Journal: Cell Rep / Year: 2016 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2n0z.cif.gz
2n0z.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2n0z.ent.gz
pdb2n0z.ent.gz PDB format
PDB format 2n0z.json.gz
2n0z.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads 2n0z_validation.pdf.gz
2n0z_validation.pdf.gz wwPDB validaton report
wwPDB validaton report 2n0z_full_validation.pdf.gz
2n0z_full_validation.pdf.gz 2n0z_validation.xml.gz
2n0z_validation.xml.gz 2n0z_validation.cif.gz
2n0z_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/n0/2n0z
https://data.pdbj.org/pub/pdb/validation_reports/n0/2n0z ftp://data.pdbj.org/pub/pdb/validation_reports/n0/2n0z
ftp://data.pdbj.org/pub/pdb/validation_reports/n0/2n0z


 Links
Links Assembly
Assembly
 Components
Components Homo sapiens (human) / Gene: MYO6 / Production host:
Homo sapiens (human) / Gene: MYO6 / Production host: 
 Sample preparation
Sample preparation Processing
Processing Movie
Movie Controller
Controller













 PDBj
PDBj








 X-PLOR NIH
X-PLOR NIH