[English] 日本語
 Yorodumi
Yorodumi- PDB-2mne: Recognition complex of DNA d(CGACTAGTCG)2 with thiazotropsin anal... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2mne | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Recognition complex of DNA d(CGACTAGTCG)2 with thiazotropsin analogue AIK-18/51 | ||||||||||||||||||||||
 Components Components | 5'-D(* Keywords KeywordsDNA / THIAZOTROPSINS / DNA RECOGNITION / ISOTHERMAL TITRATION CALORIMETRY NMR / SELF-ASSEMBLY / MINOR GROOVE BINDER | Function / homology | AIK-18/51 / DNA |  Function and homology information Function and homology informationMethod | SOLUTION NMR / molecular dynamics | Model details | minimized average structure, model 1 | Model type details | minimized average |  Authors AuthorsAlniss, H.Y. / Salvia, M.-V. / Sadikov, M. / Golovchenko, I. / Anthony, N.G. / Khalaf, A.I. / Mackay, S.P. / Suckling, C.J. / Parkinson, J.A. |  Citation Citation Journal: Chembiochem / Year: 2014 Journal: Chembiochem / Year: 2014Title: Recognition of the DNA minor groove by thiazotropsin analogues. Authors: Alniss, H.Y. / Salvia, M.V. / Sadikov, M. / Golovchenko, I. / Anthony, N.G. / Khalaf, A.I. / MacKay, S.P. / Suckling, C.J. / Parkinson, J.A. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2mne.cif.gz 2mne.cif.gz | 30.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2mne.ent.gz pdb2mne.ent.gz | 18 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2mne.json.gz 2mne.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mn/2mne https://data.pdbj.org/pub/pdb/validation_reports/mn/2mne ftp://data.pdbj.org/pub/pdb/validation_reports/mn/2mne ftp://data.pdbj.org/pub/pdb/validation_reports/mn/2mne | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2mnbC  2mndC  2mnfC C: citing same article ( |
|---|---|
| Similar structure data | |
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 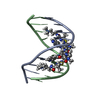
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 3045.005 Da / Num. of mol.: 2 / Source method: obtained synthetically #2: Chemical | |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 2 mM AIK-18/51, 2 mM 5'-D(*CP*GP*AP*CP*TP*AP*GP*TP*CP*G)-3', 90% H2O/10% D2O Solvent system: 90% H2O/10% D2O | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| |||||||||
| Sample conditions | Ionic strength: 0.05 / pH: 7.4 / Pressure: ambient / Temperature: 298 K |
-NMR measurement
| NMR spectrometer | Type: Bruker Avance / Manufacturer: Bruker / Model: AVANCE / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: molecular dynamics / Software ordinal: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||
| NMR constraints | NOE constraints total: 756 / NOE intraresidue total count: 336 / NOE sequential total count: 123 | ||||||||||||||||||||||||||||||||||||||||||||||||
| NMR representative | Selection criteria: minimized average structure | ||||||||||||||||||||||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the least restraint violations Conformers calculated total number: 1 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller








 PDBj
PDBj


 Amber
Amber