+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2lyg | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Fuc_TBA | |||||||||||||||||||||
 Components Components | DNA (5'-D(P* Keywords KeywordsDNA / Carbohydrate-DNA interaction / G-Quadruplex structure | Function / homology | 2-hydroxyethyl 6-deoxy-beta-L-galactopyranoside / DNA / DNA (> 10) |  Function and homology information Function and homology informationMethod | SOLUTION NMR / molecular dynamics |  Authors AuthorsGomez-Pinto, I. / Vengut-Climent, E. / Lucas, R. / Avio, A. / Eritja, R. / Gonzalez-Ibaez, C. / Morales, J. |  Citation Citation Journal: Chemistry / Year: 2013 Journal: Chemistry / Year: 2013Title: Carbohydrate-DNA interactions at G-quadruplexes: folding and stability changes by attaching sugars at the 5'-end. Authors: Gomez-Pinto, I. / Vengut-Climent, E. / Lucas, R. / Avino, A. / Eritja, R. / Gonzalez, C. / Morales, J.C. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2lyg.cif.gz 2lyg.cif.gz | 125.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2lyg.ent.gz pdb2lyg.ent.gz | 101.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2lyg.json.gz 2lyg.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2lyg_validation.pdf.gz 2lyg_validation.pdf.gz | 355 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2lyg_full_validation.pdf.gz 2lyg_full_validation.pdf.gz | 423.1 KB | Display | |
| Data in XML |  2lyg_validation.xml.gz 2lyg_validation.xml.gz | 9 KB | Display | |
| Data in CIF |  2lyg_validation.cif.gz 2lyg_validation.cif.gz | 13.5 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ly/2lyg https://data.pdbj.org/pub/pdb/validation_reports/ly/2lyg ftp://data.pdbj.org/pub/pdb/validation_reports/ly/2lyg ftp://data.pdbj.org/pub/pdb/validation_reports/ly/2lyg | HTTPS FTP |
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 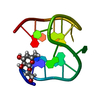
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 4743.051 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|---|
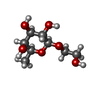
| #2: Sugar | ChemComp-1CF / |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||
| NMR details | Text: The structure was determined using NOE data |
- Sample preparation
Sample preparation
| Details | Contents: 1 mM DNA (5'-D(P*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), 1 mM SUGAR (ALPHA-D-FUCOSE), 90% H2O/10% D2O Solvent system: 90% H2O/10% D2O | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| |||||||||
| Sample conditions | pH: 7 / Pressure: ambient / Temperature: 279.6 K |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software | Name:  Amber AmberDeveloper: Case, Darden, Cheatham, III, Simmerling, Wang, Duke, Luo, ... and Kollman Classification: refinement |
|---|---|
| Refinement | Method: molecular dynamics / Software ordinal: 1 |
| NMR representative | Selection criteria: lowest energy |
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 12 / Conformers submitted total number: 12 |
 Movie
Movie Controller
Controller









 PDBj
PDBj