[English] 日本語
 Yorodumi
Yorodumi- PDB-2kf8: Structure of a two-G-tetrad basket-type intramolecular G-quadrupl... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2kf8 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of a two-G-tetrad basket-type intramolecular G-quadruplex formed by human telomeric repeats in K+ solution | ||||||
 Components Components | HUMAN TELOMERE DNA | ||||||
 Keywords Keywords | DNA / anticancer targets / human telomere / intramolecular G-quadruplexes | ||||||
| Function / homology | DNA / DNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / molecular dynamics | ||||||
| Model details | lowest energy, model 1 | ||||||
 Authors Authors | Lim, K.W. / Amrane, S. / Bouaziz, S. / Xu, W. / Mu, Y. / Patel, D.J. / Luu, K.N. / Phan, A.T. | ||||||
 Citation Citation |  Journal: J.Am.Chem.Soc. / Year: 2009 Journal: J.Am.Chem.Soc. / Year: 2009Title: Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers Authors: Lim, K.W. / Amrane, S. / Bouaziz, S. / Xu, W. / Mu, Y. / Patel, D.J. / Luu, K.N. / Phan, A.T. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2kf8.cif.gz 2kf8.cif.gz | 150.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2kf8.ent.gz pdb2kf8.ent.gz | 124.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2kf8.json.gz 2kf8.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/kf/2kf8 https://data.pdbj.org/pub/pdb/validation_reports/kf/2kf8 ftp://data.pdbj.org/pub/pdb/validation_reports/kf/2kf8 ftp://data.pdbj.org/pub/pdb/validation_reports/kf/2kf8 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 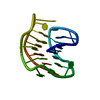
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 6974.483 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: Nucleotide synthesis |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| |||||||||||||||||||||
| Sample conditions | pH: 7 / Temperature: 298 K |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: molecular dynamics / Software ordinal: 1 Details: This natural sequence exhibits the same conformation as the 7BRG-substituted sequence, which has a much cleaner NMR spectrum. 10 relaxation matrix intensity-refined structures of the 7BRG ...Details: This natural sequence exhibits the same conformation as the 7BRG-substituted sequence, which has a much cleaner NMR spectrum. 10 relaxation matrix intensity-refined structures of the 7BRG form with the lowest energy were taken, the bromine atoms were replaced with hydrogen atoms, and the structures were subsequently subjected to distance-retrained molecular dynamics refinement. | ||||||||||||||||||||
| NMR constraints | NA alpha-angle constraints total count: 0 / NA beta-angle constraints total count: 0 / NA chi-angle constraints total count: 8 / NA delta-angle constraints total count: 0 / NA epsilon-angle constraints total count: 4 / NA gamma-angle constraints total count: 0 / NA other-angle constraints total count: 8 / NA sugar pucker constraints total count: 0 / Hydrogen bond constraints total count: 42 | ||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: all calculated structures submitted Conformers calculated total number: 10 / Conformers submitted total number: 10 / Representative conformer: 1 |
 Movie
Movie Controller
Controller
















 PDBj
PDBj






































 HSQC
HSQC