[English] 日本語
 Yorodumi
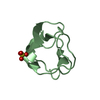
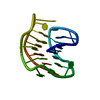
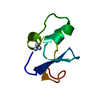
Yorodumi- PDB-1uoy: The bubble protein from Penicillium brevicompactum Dierckx exudate. -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1uoy | ||||||
|---|---|---|---|---|---|---|---|
| Title | The bubble protein from Penicillium brevicompactum Dierckx exudate. | ||||||
 Components Components | BUBBLE PROTEIN | ||||||
 Keywords Keywords | EXUDATE PROTEIN / SULFUR PHASING / POTENTIAL KILLER TOXIN | ||||||
| Function / homology | Archaeosine Trna-guanine Transglycosylase; Chain: A, domain 4 - #50 / Bubble protein / Bubble superfamily / Bubble protein / Archaeosine Trna-guanine Transglycosylase; Chain: A, domain 4 / Roll / extracellular region / Mainly Beta / Bubble protein Function and homology information Function and homology information | ||||||
| Biological species |  PENICILLIUM BREVICOMPACTUM (fungus) PENICILLIUM BREVICOMPACTUM (fungus) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SAD / Resolution: 1.5 Å SAD / Resolution: 1.5 Å | ||||||
 Authors Authors | Olsen, J.G. / Flensburg, C. / Olsen, O. / Seibold, M. / Bricogne, G. / Henriksen, A. | ||||||
 Citation Citation |  Journal: Acta Crystallogr.,Sect.D / Year: 2004 Journal: Acta Crystallogr.,Sect.D / Year: 2004Title: Solving the Structure of the Bubble Protein Using the Anomalous Sulfur Signal from Single-Crystal in-House Cu Kalpha Diffraction Data Only Authors: Olsen, J.G. / Flensburg, C. / Olsen, O. / Bricogne, G. / Henriksen, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1uoy.cif.gz 1uoy.cif.gz | 25.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1uoy.ent.gz pdb1uoy.ent.gz | 16.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1uoy.json.gz 1uoy.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1uoy_validation.pdf.gz 1uoy_validation.pdf.gz | 405.7 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1uoy_full_validation.pdf.gz 1uoy_full_validation.pdf.gz | 405.7 KB | Display | |
| Data in XML |  1uoy_validation.xml.gz 1uoy_validation.xml.gz | 6 KB | Display | |
| Data in CIF |  1uoy_validation.cif.gz 1uoy_validation.cif.gz | 8.1 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/uo/1uoy https://data.pdbj.org/pub/pdb/validation_reports/uo/1uoy ftp://data.pdbj.org/pub/pdb/validation_reports/uo/1uoy ftp://data.pdbj.org/pub/pdb/validation_reports/uo/1uoy | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 6579.132 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  PENICILLIUM BREVICOMPACTUM (fungus) / Variant: DIERCKX / References: UniProt: P83799*PLUS PENICILLIUM BREVICOMPACTUM (fungus) / Variant: DIERCKX / References: UniProt: P83799*PLUS |
|---|---|
| #2: Water | ChemComp-HOH / |
| Has protein modification | Y |
| Sequence details | SEQUENCE YET TO BE DEPOSITED IN PUBLIC DATABASE |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.6 Å3/Da / Density % sol: 52 % Description: PHASING USING THE ANOMALOUS SIGNAL FROM 8 CYSTEINE SULFURS (4 DISULFIDE BRIDGES). THE IMAGE PLATE MODEL USED WAS RAXIS IV++ | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 277 K / pH: 5 Details: 16 MG/ML PROTEIN IN 30 MM NA-ACETATE BUFFER, PH 5.0 AT 4 DEG. | |||||||||||||||
| Crystal grow | *PLUS Temperature: 277 K / Method: vapor diffusion, hanging drop | |||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 120 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RUH3R / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RUH3R / Wavelength: 1.5418 |
| Detector | Type: RIGAKU IMAGE PLATE / Detector: IMAGE PLATE / Date: Jan 15, 2002 / Details: MAX-FLUX OPTICAL SYSTEM |
| Radiation | Monochromator: MIRRORS / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.5→15.2 Å / Num. obs: 11250 / % possible obs: 99.9 % / Redundancy: 13.5 % / Biso Wilson estimate: 14 Å2 / Rmerge(I) obs: 0.055 / Net I/σ(I): 8.3 |
| Reflection shell | Resolution: 1.5→1.58 Å / Redundancy: 12.3 % / Rmerge(I) obs: 0.155 / Mean I/σ(I) obs: 3.6 / % possible all: 100 |
| Reflection | *PLUS Highest resolution: 1.5 Å / Lowest resolution: 15 Å / Redundancy: 13.5 % / Num. measured all: 151997 / Rmerge(I) obs: 0.055 |
| Reflection shell | *PLUS % possible obs: 99.9 % / Rmerge(I) obs: 0.155 / Mean I/σ(I) obs: 3.6 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  SAD / Resolution: 1.5→15.21 Å / Rfactor Rfree error: 0.006 / Data cutoff high absF: 834780.52 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 SAD / Resolution: 1.5→15.21 Å / Rfactor Rfree error: 0.006 / Data cutoff high absF: 834780.52 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 37.1916 Å2 / ksol: 0.340968 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 14.7 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.5→15.21 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.5→1.59 Å / Rfactor Rfree error: 0.019 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Lowest resolution: 15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller







 PDBj
PDBj
