[English] 日本語
 Yorodumi
Yorodumi- PDB-2jsl: Monomeric Human Telomere DNA Tetraplex with 3+1 Strand Fold Topol... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2jsl | ||||||
|---|---|---|---|---|---|---|---|
| Title | Monomeric Human Telomere DNA Tetraplex with 3+1 Strand Fold Topology, Two Edgewise Loops and Double-Chain Reversal Loop, Form 2 Natural, NMR, 10 Structures | ||||||
 Components Components | HUMAN TELOMERE DNA | ||||||
 Keywords Keywords | DNA / G-TETRAD / 3+1 STRAND FOLDING / EDGEWISE / DOUBLE-CHAIN REVERSAL LOOPS / 10 STRUCTURES / Form 2 Natural | ||||||
| Function / homology | DNA / DNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / TORSION ANGLE DYNAMICS CARTESIAN DYNAMICS MATRIX RELAXATION | ||||||
 Authors Authors | Kuryavyi, V.V. / Phan, A.T. / Luu, K.N. / Patel, D.J. | ||||||
 Citation Citation |  Journal: Nucleic Acids Res. / Year: 2007 Journal: Nucleic Acids Res. / Year: 2007Title: Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Authors: Phan, A.T. / Kuryavyi, V. / Luu, K.N. / Patel, D.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2jsl.cif.gz 2jsl.cif.gz | 157.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2jsl.ent.gz pdb2jsl.ent.gz | 130.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2jsl.json.gz 2jsl.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/js/2jsl https://data.pdbj.org/pub/pdb/validation_reports/js/2jsl ftp://data.pdbj.org/pub/pdb/validation_reports/js/2jsl ftp://data.pdbj.org/pub/pdb/validation_reports/js/2jsl | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 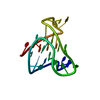
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 7896.076 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 0.5-5.0 mM HUMAN TELOMERE DNA, 100% D2O / Solvent system: 100% D2O |
|---|---|
| Sample | Conc.: 0.5 mM / Component: HUMAN TELOMERE DNA |
| Sample conditions | Ionic strength: 70 / pH: 7 / Pressure: 1 atm / Temperature: 298 K |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: TORSION ANGLE DYNAMICS CARTESIAN DYNAMICS MATRIX RELAXATION Software ordinal: 1 Details: AFTER DEPOSITION, THE MOLECULES WERE ENERGY MINIMIZED WITH THE ENERGY FUNCTION IMPLEMENTING GEOMETRICAL VALUES IN EFFECT AT RCSB SINCE JULY 31 2007. AS A RESULT, STRUCTURE STATISTICS FOR ...Details: AFTER DEPOSITION, THE MOLECULES WERE ENERGY MINIMIZED WITH THE ENERGY FUNCTION IMPLEMENTING GEOMETRICAL VALUES IN EFFECT AT RCSB SINCE JULY 31 2007. AS A RESULT, STRUCTURE STATISTICS FOR THIS ENTRY SLIGHTLY DEVIATES FROM THE PUBLISHED ONE, AS FOLLOWS (CURRENT VS PUBLISHED): NOE VIOLATIONS (0.7+-0.67) VS (0.30+-0.48); MAXIMUM VIOLATION (0.23+-0.03) VS (0.23+-0.02); RMSD OF VIOLATIONS (0.04+-0.00) VS (0.03+-0.00); BOND LENGTHS (0.005+-0.000) VS (0.004+-0.000); BOND ANGLES (0.77+-0.03) VS (0.87+-0.01); IMPROPERS (0.41+-0.02) VS (0.37+-0.02); PAIRWISE RMSD: ALL ATOMS (1.26+-0.31) VS (1.46+-0.36); ALL ATOMS EXCEPT T18, T19, A20 (0.73+-0.12) VS (0.79+-0.13). | ||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 50 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller















 PDBj
PDBj






































 X-PLOR
X-PLOR