[English] 日本語
 Yorodumi
Yorodumi- PDB-2kcx: Solution NMR Structure of Kazal-1 Domain of Human Follistatin-rel... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2kcx | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution NMR Structure of Kazal-1 Domain of Human Follistatin-related protein 3 (FSTL-3). Northeast Structural Genomics Target HR6186A. | ||||||
 Components Components | Follistatin-related protein 3 | ||||||
 Keywords Keywords | structural genomics / unknown function / Kazal-1 / Follistatin / Chromosomal rearrangement / Glycoprotein / Nucleus / Proto-oncogene / Secreted / PSI-2 / Protein Structure Initiative / Northeast Structural Genomics Consortium / NESG / NUCLEOTIDE-BINDING PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway / Antagonism of Activin by Follistatin / regulation of BMP signaling pathway / positive regulation of cell-cell adhesion / activin binding / negative regulation of activin receptor signaling pathway / negative regulation of osteoclast differentiation / fibronectin binding / negative regulation of BMP signaling pathway / hematopoietic progenitor cell differentiation ...negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway / Antagonism of Activin by Follistatin / regulation of BMP signaling pathway / positive regulation of cell-cell adhesion / activin binding / negative regulation of activin receptor signaling pathway / negative regulation of osteoclast differentiation / fibronectin binding / negative regulation of BMP signaling pathway / hematopoietic progenitor cell differentiation / ossification / Post-translational protein phosphorylation / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / cell differentiation / endoplasmic reticulum lumen / regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / extracellular space / extracellular region / nucleoplasm / nucleus Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | SOLUTION NMR / torsion angle dynamics | ||||||
| Model details | lowest energy, model 1 | ||||||
 Authors Authors | Rossi, P. / Chiang, Y. / Anderson, S. / Montelione, G.T. / Northeast Structural Genomics Consortium (NESG) | ||||||
 Citation Citation |  Journal: To be Published Journal: To be PublishedTitle: Solution NMR Structure of Kazal-1 Domain of Human Follistatin-related protein 3 (FSTL-3). Northeast Structural Genomics Target HR6186A. Authors: Rossi, P. / Chiang, Y. / Anderson, S. / Montelione, G.T. | ||||||
| History |
|
- Structure visualization
Structure visualization
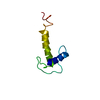
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2kcx.cif.gz 2kcx.cif.gz | 461.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2kcx.ent.gz pdb2kcx.ent.gz | 390.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2kcx.json.gz 2kcx.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/kc/2kcx https://data.pdbj.org/pub/pdb/validation_reports/kc/2kcx ftp://data.pdbj.org/pub/pdb/validation_reports/kc/2kcx ftp://data.pdbj.org/pub/pdb/validation_reports/kc/2kcx | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 7981.164 Da / Num. of mol.: 1 / Fragment: Kazal-1 domain, residues 97-169 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: FSTL3, FLRG, UNQ674/PRO1308 / Production host: Homo sapiens (human) / Gene: FSTL3, FLRG, UNQ674/PRO1308 / Production host:  |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||||||||||
| Sample conditions | Ionic strength: 0.2 / pH: 6.5 / Pressure: ambient / Temperature: 298 K |
-NMR measurement
| NMR spectrometer | Type: Bruker Avance / Manufacturer: Bruker / Model: AVANCE / Field strength: 800 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: torsion angle dynamics / Software ordinal: 1 Details: MONOMER BY GEL FILTRATION CHROMATOGRAPHY/LIGHT SCATTERING AND BY NMR. T1/T2(CPMG) (MS) = 516.1/136.2, TAUC = 3.94(NS). CONSISTENT WITH MOLECULAR WEIGHT. STRUCTURE DETERMINED BY TRIPLE ...Details: MONOMER BY GEL FILTRATION CHROMATOGRAPHY/LIGHT SCATTERING AND BY NMR. T1/T2(CPMG) (MS) = 516.1/136.2, TAUC = 3.94(NS). CONSISTENT WITH MOLECULAR WEIGHT. STRUCTURE DETERMINED BY TRIPLE RESONANCE NMR SPECTROSCOPY. NOESY ASSIGNMENTS BY CYANA2.1. 20 OF 100 STRUCTURES LOWEST TARGET FUNCTION SELECTED WITH CYANA-2.1. SELECTED MODELS ARE FURTHER REFINED USING CNS IN EXPLICIT WATER SHELL (NILGES PROTOCOL WITH PARAM19). ASSIGNMENT STATS (EXCLUDING C-TERM TAG): BACKBONE 98.10%, SIDECHAIN 88.50%, AROMATIC (SC) 75.00%. STRUCTURE BASED ON 912 NOE, 65 DIHE. MAX NOE VIOLATION: 0.19 A (1MODEL); MAX DIHE VIOLATION: 5.4 DEG. 6 TOTAL CLOSE CONTACTS PER 20 MODELS. STRUCTURE QUALITY FACTOR (PSVS 1.3): ORDERED RESIDUES RANGES: 4-9, 13-27, 33-34, 37-58, 61-69, 72-73 FOR [S(PHI)+S(PSI)] > 1.8. SECONDARY STRUCTURE - ALPHA HELICES: 50-60, BETA STRANDS: 13-18, 21-26, 46-47, 39-41, 66-69. RMSD 1.3 BACKBONE, 1.8 ALL HEAVY ATOMS. RAMA. DISTRIBUTION: 92.1/7.8/0.1/0.0. PROCHECK (PSI-PHI): -0.34/-1.02 (RAW/Z), PROCHECK (ALL): -0.33/-1.95 (RAW/Z), MOLPROBITY CLASH: 19.69/-1.85 (RAW/Z). RPF SCORES ALL ASSIGNED RESIDUES (FIT OF NOESY PEAKLISTS TO STRUCTURE): RECALL: 0.99, PRECISION: 0.85, F-MEASURE: 0.91, DP-SCORE: 0.82. | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: target function / Conformers calculated total number: 100 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller









 PDBj
PDBj
 HSQC
HSQC