+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2iq9 | ||||||
|---|---|---|---|---|---|---|---|
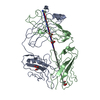
| Title | PFA2 FAB fragment, triclinic apo form | ||||||
 Components Components |
| ||||||
 Keywords Keywords | IMMUNE SYSTEM / pfa2 / wwddd / cdr | ||||||
| Function / homology |  Function and homology information Function and homology informationimmunoglobulin complex / adaptive immune response / extracellular region / metal ion binding / plasma membrane Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.3 Å MOLECULAR REPLACEMENT / Resolution: 2.3 Å | ||||||
 Authors Authors | Gardberg, A.S. / Dealwis, C. | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.Usa / Year: 2007 Journal: Proc.Natl.Acad.Sci.Usa / Year: 2007Title: Molecular basis for passive immunotherapy of Alzheimer's disease Authors: Gardberg, A.S. / Dice, L.T. / Ou, S. / Rich, R.L. / Helmbrecht, E. / Ko, J. / Wetzel, R. / Myszka, D.G. / Patterson, P.H. / Dealwis, C. | ||||||
| History |
| ||||||
| Remark 999 | SEQUENCE Presently, there is no aminoacid sequence database reference available for the two ...SEQUENCE Presently, there is no aminoacid sequence database reference available for the two proteins IGG2A FAB fragment heavy chain and IGG2A FAB fragment light chain kappa |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2iq9.cif.gz 2iq9.cif.gz | 100.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2iq9.ent.gz pdb2iq9.ent.gz | 75.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2iq9.json.gz 2iq9.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/iq/2iq9 https://data.pdbj.org/pub/pdb/validation_reports/iq/2iq9 ftp://data.pdbj.org/pub/pdb/validation_reports/iq/2iq9 ftp://data.pdbj.org/pub/pdb/validation_reports/iq/2iq9 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2iptC  2ipuC  2iqaC  2r0wC  2r0zC  1ncwS  2iqb C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | The biological assembly is a heterodimer. |
- Components
Components
| #1: Antibody | Mass: 24146.812 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: hybridoma / Source: (natural)  | ||||
|---|---|---|---|---|---|
| #2: Antibody | Mass: 23903.994 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: hybridoma / Source: (natural)  | ||||
| #3: Chemical | | #4: Water | ChemComp-HOH / | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.14 Å3/Da / Density % sol: 42.49 % |
|---|---|
| Crystal grow | Temperature: 294 K / Method: vapor diffusion / pH: 8.5 Details: 25% PEG3350, 0.1 M Tris, pH 8.5, VAPOR DIFFUSION, temperature 294K |
-Data collection
| Diffraction | Mean temperature: 113 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU / Wavelength: 1.54 Å ROTATING ANODE / Type: RIGAKU / Wavelength: 1.54 Å |
| Detector | Type: RIGAKU RAXIS IV / Detector: IMAGE PLATE / Date: Oct 12, 2005 / Details: mirrors |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.54 Å / Relative weight: 1 |
| Reflection | Resolution: 2.3→22 Å / Num. obs: 16504 / % possible obs: 93.3 % / Redundancy: 1.7 % / Biso Wilson estimate: 38.6 Å2 / Rsym value: 0.072 / Net I/σ(I): 10.8 |
| Reflection shell | Resolution: 2.3→2.38 Å / Redundancy: 1.7 % / Mean I/σ(I) obs: 2 / Num. unique all: 1639 / Rsym value: 0.359 / % possible all: 92.8 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1NCW Resolution: 2.3→20.7 Å / Cor.coef. Fo:Fc: 0.937 / Cor.coef. Fo:Fc free: 0.9 / SU B: 10.855 / SU ML: 0.26 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.79 / ESU R Free: 0.317 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 27.147 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.3→20.7 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.3→2.359 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller












 PDBj
PDBj