登録情報 データベース : PDB / ID : 2haxタイトル Crystal structure of Bacillus caldolyticus cold shock protein in complex with hexathymidine 5'-D(*TP*TP*TP*TP*TP*T)-3'Cold shock protein cspB キーワード / / / / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
/ / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Bacillus caldolyticus (バクテリア)手法 / / / 解像度 : 1.29 Å データ登録者 Max, K.E.A. / Heinemann, U. ジャーナル : Febs J. / 年 : 2007タイトル : Common mode of DNA binding to cold shock domains. Crystal structure of hexathymidine bound to the domain-swapped form of a major cold shock protein from Bacillus caldolyticus.著者 : Max, K.E. / Zeeb, M. / Bienert, R. / Balbach, J. / Heinemann, U. 履歴 登録 2006年6月13日 登録サイト / 処理サイト 改定 1.0 2007年4月24日 Provider / タイプ 改定 1.1 2008年4月1日 Group 改定 1.2 2011年7月13日 Group 改定 1.3 2017年10月18日 Group / カテゴリ / Item / _software.name改定 1.4 2023年8月30日 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Refinement description カテゴリ chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / pdbx_struct_conn_angle / struct_conn / struct_site Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_symmetry / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_symmetry / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_conn.ptnr2_symmetry / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id
すべて表示 表示を減らす Remark 300 BIOMOLECULE: 1 THIS ENTRY CONTAINS THE CRYSTALLOGRAPHIC ASYMMETRIC UNIT WHICH CONSISTS OF 4 ... BIOMOLECULE: 1 THIS ENTRY CONTAINS THE CRYSTALLOGRAPHIC ASYMMETRIC UNIT WHICH CONSISTS OF 4 CHAIN(S). SEE REMARK 350 FOR INFORMATION ON GENERATING THE BIOLOGICAL MOLECULE(S). THE AUTHORS STATE THAT THE STRUCTURE SHOWS A SWAPPED PROTEIN DIMER (2 CHAINS) IN COMPLEX WITH TWO HEXANUCLEOTIDES, WHEREAS IN SOLUTION THEY ONLY OBSERVE A SINGLE PROTEIN CHAIN IN COMPLEX WITH A SINGLE HEXANUCLEOTIDE. THEY THEREFORE EXPECT THAT THE BIOLOGICAL UNIT IS A SINGLE CLOSED MONOMERIC PROTEIN CHAIN WITH A SINGLE LIGAND MOLECULE. SUCH A CLOSED MONOMERIC ENTITY WOULD CORRESPOND TO RESIDUES 1-36 OF CHAIN A AND RESIDUES 37-66 OF CHAIN B AND IT WOULD BE IN COMPLEX WITH LIGAND MOLECULE C. A SECOND BIOLOGICAL UNIT WOULD CONSIST OF RESIDUES 1-36 OF CHAIN B AND RESIDUES 37-66 OF CHAIN A. THIS MONOMERIC ENTITY WOULD BE IN COMPLEX WITH MOLECULE D.
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Bacillus caldolyticus (バクテリア)
Bacillus caldolyticus (バクテリア) X線回折 /
X線回折 /  シンクロトロン /
シンクロトロン /  分子置換 / 解像度: 1.29 Å
分子置換 / 解像度: 1.29 Å  データ登録者
データ登録者 引用
引用 ジャーナル: Febs J. / 年: 2007
ジャーナル: Febs J. / 年: 2007 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 2hax.cif.gz
2hax.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb2hax.ent.gz
pdb2hax.ent.gz PDB形式
PDB形式 2hax.json.gz
2hax.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード https://data.pdbj.org/pub/pdb/validation_reports/ha/2hax
https://data.pdbj.org/pub/pdb/validation_reports/ha/2hax ftp://data.pdbj.org/pub/pdb/validation_reports/ha/2hax
ftp://data.pdbj.org/pub/pdb/validation_reports/ha/2hax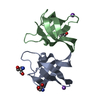
 リンク
リンク 集合体
集合体
 要素
要素 Bacillus caldolyticus (バクテリア)
Bacillus caldolyticus (バクテリア)
 X線回折 / 使用した結晶の数: 1
X線回折 / 使用した結晶の数: 1  試料調製
試料調製 シンクロトロン / サイト:
シンクロトロン / サイト:  BESSY
BESSY  / ビームライン: 14.1 / 波長: 0.9184 Å
/ ビームライン: 14.1 / 波長: 0.9184 Å 解析
解析 分子置換
分子置換 ムービー
ムービー コントローラー
コントローラー
















 PDBj
PDBj




