+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2h89 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Avian Respiratory Complex II with Malonate Bound | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | OXIDOREDUCTASE / COMPLEX II / MEMBRANE PROTEIN / HEME PROTEIN / IRON SULFUR PROTEIN / CYTOCHROME B / REDOX ENZYME / RESPIRATORY CHAIN / MALONATE / UBIQUINONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationThe tricarboxylic acid cycle / Oxidoreductases; Acting on the CH-OH group of donors; With a quinone or similar compound as acceptor / succinate metabolic process / respiratory chain complex II (succinate dehydrogenase) / mitochondrial electron transport, succinate to ubiquinone / succinate dehydrogenase (quinone) activity / succinate dehydrogenase / 3 iron, 4 sulfur cluster binding / ubiquinone binding / tricarboxylic acid cycle ...The tricarboxylic acid cycle / Oxidoreductases; Acting on the CH-OH group of donors; With a quinone or similar compound as acceptor / succinate metabolic process / respiratory chain complex II (succinate dehydrogenase) / mitochondrial electron transport, succinate to ubiquinone / succinate dehydrogenase (quinone) activity / succinate dehydrogenase / 3 iron, 4 sulfur cluster binding / ubiquinone binding / tricarboxylic acid cycle / aerobic respiration / respiratory electron transport chain / 2 iron, 2 sulfur cluster binding / mitochondrial membrane / flavin adenine dinucleotide binding / 4 iron, 4 sulfur cluster binding / electron transfer activity / mitochondrial inner membrane / heme binding / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / rigid body refinement of previous / Resolution: 2.4 Å SYNCHROTRON / rigid body refinement of previous / Resolution: 2.4 Å | |||||||||
 Authors Authors | Huang, L.S. / Shen, J.T. / Wang, A.C. / Berry, E.A. | |||||||||
 Citation Citation |  Journal: Biochim.Biophys.Acta Journal: Biochim.Biophys.ActaTitle: Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the Authors: Huang, L.S. / Shen, J.T. / Wang, A.C. / Berry, E.A. #1:  Journal: J.Biol.Chem. / Year: 2006 Journal: J.Biol.Chem. / Year: 2006Title: 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme. Authors: Huang, L.S. / Sun, G. / Cobessi, D. / Wang, A.C. / Shen, J.T. / Tung, E.Y. / Anderson, V.E. / Berry, E.A. #2:  Journal: Acta Crystallogr.,Sect.D / Year: 2005 Journal: Acta Crystallogr.,Sect.D / Year: 2005Title: Crystallization of mitochondrial respiratory complex II from chicken heart: a membrane-protein complex diffracting to 2.0 A Authors: Huang, L.S. / Borders, T.M. / Shen, J.T. / Wang, C.J. / Berry, E.A. #3:  Journal: Cell(Cambridge,Mass.) / Year: 2005 Journal: Cell(Cambridge,Mass.) / Year: 2005Title: Crystal structure of mitochondrial respiratory membrane protein complex II. Authors: Sun, F. / Huo, X. / Zhai, Y. / Wang, A. / Xu, J. / Su, D. / Bartlam, M. / Rao, Z. | |||||||||
| History |
| |||||||||
| Remark 999 | SEQUENCE SEQUENCE 1-71 OF SUCCINATE DEHYDROGENASE FP SUBUNIT DOES NOT MATCH TO ANY SEQUENCE IN THE ...SEQUENCE SEQUENCE 1-71 OF SUCCINATE DEHYDROGENASE FP SUBUNIT DOES NOT MATCH TO ANY SEQUENCE IN THE DATABASE. THE SEQUENCE OF SUCCINATE DEHYDROGENASE CYTOCHROME B, LARGE SUBUNIT IS NOT AVAILABLE IN ANY OF THE DATABASE SEQUENCES AT THE TIME OF PROCESSING. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2h89.cif.gz 2h89.cif.gz | 255.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2h89.ent.gz pdb2h89.ent.gz | 196.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2h89.json.gz 2h89.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/h8/2h89 https://data.pdbj.org/pub/pdb/validation_reports/h8/2h89 ftp://data.pdbj.org/pub/pdb/validation_reports/h8/2h89 ftp://data.pdbj.org/pub/pdb/validation_reports/h8/2h89 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2h88C  1yq3S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 2 types, 2 molecules AB
| #1: Protein | Mass: 68256.922 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #2: Protein | Mass: 28685.221 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-SUCCINATE DEHYDROGENASE CYTOCHROME B, ... , 2 types, 2 molecules CD
| #3: Protein | Mass: 15392.052 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #4: Protein | Mass: 10971.604 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Sugars , 1 types, 1 molecules 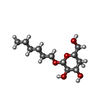
| #12: Sugar | ChemComp-BHG / |
|---|
-Non-polymers , 10 types, 630 molecules 
















| #5: Chemical | | #6: Chemical | ChemComp-FAD / | #7: Chemical | ChemComp-MLI / | #8: Chemical | ChemComp-UNL / Num. of mol.: 40 / Source method: obtained synthetically #9: Chemical | ChemComp-FES / | #10: Chemical | ChemComp-SF4 / | #11: Chemical | ChemComp-F3S / | #13: Chemical | ChemComp-HEM / | #14: Chemical | #15: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.51 Å3/Da / Density % sol: 64.99 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, sitting drop / pH: 7.5 Details: 50 G/L PEG-3350, 25 ML/L ISOPROPANOL, 15 ML/L PEG-400 0.05 M NA-HEPES, 0.01 M TRIS-HCL, 0.0005 M MALONATE, 0.0005 M MNCL2, 0.0013 M MGCL2, 0.0015 M NA-AZIDE, 0.00025 M NA-EDTA., pH 7.5, ...Details: 50 G/L PEG-3350, 25 ML/L ISOPROPANOL, 15 ML/L PEG-400 0.05 M NA-HEPES, 0.01 M TRIS-HCL, 0.0005 M MALONATE, 0.0005 M MNCL2, 0.0013 M MGCL2, 0.0015 M NA-AZIDE, 0.00025 M NA-EDTA., pH 7.5, VAPOR DIFFUSION, SITTING DROP, temperature 277K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 5.0.2 / Wavelength: 1.1 Å / Beamline: 5.0.2 / Wavelength: 1.1 Å |
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Jul 25, 2004 |
| Radiation | Monochromator: Double-crystal, Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.29→60 Å / Num. all: 63148 / Num. obs: 63148 / % possible obs: 78.6 % / Observed criterion σ(F): -999 / Observed criterion σ(I): -3 / Redundancy: 2.4 % / Biso Wilson estimate: 52.7 Å2 / Rsym value: 0.158 / Net I/σ(I): 7 |
| Reflection shell | Resolution: 2.29→2.33 Å / Redundancy: 1.5 % / Mean I/σ(I) obs: 1.2 / Num. unique all: 724 / Rsym value: 0.545 / % possible all: 18.5 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: rigid body refinement of previous Starting model: 1YQ3 Resolution: 2.4→50.74 Å / Rfactor Rfree error: 0.005 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 60.4229 Å2 / ksol: 0.257581 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 59.8 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.4→50.74 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.4→2.46 Å / Rfactor Rfree error: 0.037 / Total num. of bins used: 15
|
 Movie
Movie Controller
Controller



























 PDBj
PDBj






















