Entry Database : PDB / ID : 2fbwTitle Avian respiratory complex II with carboxin bound (Succinate dehydrogenase [ubiquinone] ...) x 3 Succinate dehydrogenase cytochrome b560 subunit, mitochondrial Keywords / / / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Gallus gallus (chicken)Method / / / Resolution : 2.06 Å Authors Huang, L.S. / Sun, G. / Cobessi, D. / Wang, A.C. / Shen, J.T. / Tung, E.Y. / Anderson, V.E. / Berry, E.A. Funding support Organization Grant number Country National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) R01GM62563
Journal : J.Biol.Chem. / Year : 2006Title : 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme.
Authors :
Huang, L.S. / Sun, G. / Cobessi, D. / Wang, A.C. / Shen, J.T. / Tung, E.Y. / Anderson, V.E. / Berry, E.A. History Deposition Dec 10, 2005 Deposition site / Processing site Revision 1.0 Dec 20, 2005 Provider / Type Revision 1.1 May 1, 2008 Group Revision 1.2 Jul 13, 2011 Group / Version format complianceRevision 1.3 Oct 29, 2014 Group Revision 1.4 Jul 29, 2020 Group / Derived calculations / Structure summaryCategory chem_comp / database_PDB_caveat ... chem_comp / database_PDB_caveat / entity / pdbx_entity_nonpoly / pdbx_struct_conn_angle / struct_conn / struct_site / struct_site_gen Item _chem_comp.mon_nstd_flag / _chem_comp.name ... _chem_comp.mon_nstd_flag / _chem_comp.name / _chem_comp.type / _entity.pdbx_description / _pdbx_entity_nonpoly.name / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr2_auth_asym_id / _pdbx_struct_conn_angle.ptnr2_auth_comp_id / _pdbx_struct_conn_angle.ptnr2_auth_seq_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr2_label_atom_id / _pdbx_struct_conn_angle.ptnr2_label_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.conn_type_id / _struct_conn.id / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id Description / Provider / Type Revision 2.0 Feb 17, 2021 Group Advisory / Atomic model ... Advisory / Atomic model / Author supporting evidence / Data collection / Database references / Derived calculations / Non-polymer description / Other / Polymer sequence / Refinement description / Source and taxonomy / Structure summary Category atom_site / atom_site_anisotrop ... atom_site / atom_site_anisotrop / atom_type / audit_author / cell / chem_comp / database_PDB_caveat / diffrn / diffrn_source / entity / entity_name_com / entity_poly / entity_poly_seq / entity_src_nat / pdbx_audit_support / pdbx_chem_comp_identifier / pdbx_database_related / pdbx_database_remark / pdbx_distant_solvent_atoms / pdbx_entity_instance_feature / pdbx_entity_nonpoly / pdbx_entry_details / pdbx_nonpoly_scheme / pdbx_poly_seq_scheme / pdbx_refine_tls / pdbx_refine_tls_group / pdbx_struct_assembly_gen / pdbx_struct_conn_angle / pdbx_struct_sheet_hbond / pdbx_unobs_or_zero_occ_atoms / pdbx_unobs_or_zero_occ_residues / pdbx_validate_chiral / pdbx_validate_close_contact / pdbx_validate_planes / pdbx_validate_rmsd_angle / pdbx_validate_torsion / pdbx_xplor_file / refine / refine_analyze / refine_hist / refine_ls_restr / refine_ls_shell / reflns / reflns_shell / software / struct / struct_asym / struct_biol / struct_conf / struct_conn / struct_mon_prot_cis / struct_ncs_dom_lim / struct_ref / struct_ref_seq / struct_sheet / struct_sheet_order / struct_sheet_range / symmetry Item _audit_author.identifier_ORCID / _cell.volume ... _audit_author.identifier_ORCID / _cell.volume / _chem_comp.formula / _chem_comp.formula_weight / _chem_comp.id / _chem_comp.mon_nstd_flag / _chem_comp.name / _chem_comp.pdbx_synonyms / _chem_comp.type / _diffrn.pdbx_serial_crystal_experiment / _diffrn_source.pdbx_wavelength / _entity_poly.pdbx_seq_one_letter_code / _entity_poly.pdbx_seq_one_letter_code_can / _entity_src_nat.common_name / _entity_src_nat.pdbx_beg_seq_num / _entity_src_nat.pdbx_end_seq_num / _pdbx_struct_assembly_gen.asym_id_list / _refine.B_iso_mean / _refine.aniso_B[1][1] / _refine.aniso_B[1][2] / _refine.aniso_B[1][3] / _refine.aniso_B[2][2] / _refine.aniso_B[2][3] / _refine.aniso_B[3][3] / _refine.details / _refine.ls_R_factor_R_free / _refine.ls_R_factor_R_free_error / _refine.ls_R_factor_R_work / _refine.ls_R_factor_obs / _refine.ls_d_res_high / _refine.ls_number_reflns_R_free / _refine.ls_number_reflns_R_work / _refine.ls_number_reflns_all / _refine.ls_number_reflns_obs / _refine.ls_percent_reflns_R_free / _refine.ls_percent_reflns_obs / _refine.overall_SU_ML / _refine.pdbx_data_cutoff_high_absF / _refine.pdbx_data_cutoff_low_absF / _refine.pdbx_isotropic_thermal_model / _refine.pdbx_ls_sigma_F / _refine.pdbx_overall_phase_error / _refine.pdbx_solvent_shrinkage_radii / _refine.pdbx_solvent_vdw_probe_radii / _refine.pdbx_stereochemistry_target_values / _refine.solvent_model_details / _refine.solvent_model_param_bsol / _refine.solvent_model_param_ksol / _refine_hist.d_res_high / _refine_hist.number_atoms_solvent / _refine_hist.number_atoms_total / _refine_hist.pdbx_number_atoms_ligand / _refine_hist.pdbx_number_atoms_protein / _reflns.B_iso_Wilson_estimate / _reflns.d_resolution_high / _reflns.d_resolution_low / _reflns.number_all / _reflns.number_obs / _reflns.pdbx_CC_half / _reflns.pdbx_CC_star / _reflns.pdbx_Rmerge_I_obs / _reflns.pdbx_Rpim_I_all / _reflns.pdbx_Rrim_I_all / _reflns.pdbx_Rsym_value / _reflns.pdbx_netI_over_sigmaI / _reflns.pdbx_redundancy / _reflns.percent_possible_obs / _reflns_shell.Rmerge_I_obs / _reflns_shell.d_res_high / _reflns_shell.d_res_low / _reflns_shell.meanI_over_sigI_obs / _reflns_shell.number_unique_obs / _reflns_shell.pdbx_CC_half / _reflns_shell.pdbx_Rpim_I_all / _reflns_shell.pdbx_Rrim_I_all / _reflns_shell.pdbx_Rsym_value / _reflns_shell.pdbx_redundancy / _software.classification / _software.name / _software.version / _struct.pdbx_CASP_flag / _struct_conf.beg_auth_comp_id / _struct_conf.beg_auth_seq_id / _struct_conf.beg_label_comp_id / _struct_conf.beg_label_seq_id / _struct_conf.end_auth_comp_id / _struct_conf.end_auth_seq_id / _struct_conf.end_label_comp_id / _struct_conf.end_label_seq_id / _struct_conf.pdbx_PDB_helix_id / _struct_conf.pdbx_PDB_helix_length / _struct_mon_prot_cis.pdbx_omega_angle / _struct_ref.db_code / _struct_ref.db_name / _struct_ref.entity_id / _struct_ref.pdbx_align_begin / _struct_ref.pdbx_db_accession / _struct_ref.pdbx_seq_one_letter_code / _struct_ref_seq.db_align_beg / _struct_ref_seq.db_align_end / _struct_ref_seq.pdbx_auth_seq_align_beg / _struct_ref_seq.pdbx_auth_seq_align_end / _struct_ref_seq.pdbx_db_accession / _struct_ref_seq.pdbx_strand_id / _struct_ref_seq.ref_id / _struct_ref_seq.seq_align_beg / _struct_ref_seq.seq_align_end / _struct_sheet.id / _struct_sheet.number_strands / _symmetry.space_group_name_Hall Description Details Provider / Type Revision 2.1 Aug 30, 2023 Group Advisory / Data collection ... Advisory / Data collection / Database references / Refinement description Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / pdbx_unobs_or_zero_occ_atoms Item / _database_2.pdbx_database_accessionRevision 2.2 Nov 13, 2024 Group / Category / pdbx_modification_feature / Item
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.06 Å
MOLECULAR REPLACEMENT / Resolution: 2.06 Å  Authors
Authors United States, 1items
United States, 1items  Citation
Citation Journal: J.Biol.Chem. / Year: 2006
Journal: J.Biol.Chem. / Year: 2006 Journal: Acta Crystallogr.,Sect.D / Year: 2005
Journal: Acta Crystallogr.,Sect.D / Year: 2005 Journal: Cell(Cambridge,Mass.) / Year: 2005
Journal: Cell(Cambridge,Mass.) / Year: 2005 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2fbw.cif.gz
2fbw.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2fbw.ent.gz
pdb2fbw.ent.gz PDB format
PDB format 2fbw.json.gz
2fbw.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/fb/2fbw
https://data.pdbj.org/pub/pdb/validation_reports/fb/2fbw ftp://data.pdbj.org/pub/pdb/validation_reports/fb/2fbw
ftp://data.pdbj.org/pub/pdb/validation_reports/fb/2fbw


 Links
Links Assembly
Assembly


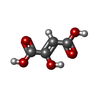
 Components
Components





























 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ALS
ALS  / Beamline: 8.2.2 / Wavelength: 0.98 Å
/ Beamline: 8.2.2 / Wavelength: 0.98 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller

















 PDBj
PDBj

























