+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2erl | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | PHEROMONE ER-1 FROM | |||||||||
 Components Components | MATING PHEROMONE ER-1 | |||||||||
 Keywords Keywords | PHEROMONE | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Euplotes raikovi (eukaryote) Euplotes raikovi (eukaryote) | |||||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1 Å X-RAY DIFFRACTION / Resolution: 1 Å | |||||||||
 Authors Authors | Anderson, D.H. / Weiss, M.S. / Eisenberg, D. | |||||||||
 Citation Citation |  Journal: Acta Crystallogr.,Sect.D / Year: 1996 Journal: Acta Crystallogr.,Sect.D / Year: 1996Title: A challenging case for protein crystal structure determination: the mating pheromone Er-1 from Euplotes raikovi. Authors: Anderson, D.H. / Weiss, M.S. / Eisenberg, D. #1:  Journal: Proc.Natl.Acad.Sci.USA / Year: 1995 Journal: Proc.Natl.Acad.Sci.USA / Year: 1995Title: A Cooperative Model for Receptor Recognition and Cell Adhesion: Evidence from the Molecular Packing in the 1.6-A Crystal Structure of the Pheromone Er-1 from the Ciliated Protozoan Euplotes Raikovi Authors: Weiss, M.S. / Anderson, D.H. / Raffioni, S. / Bradshaw, R.A. / Ortenzi, C. / Luporini, P. / Eisenberg, D. #2:  Journal: Protein Sci. / Year: 1994 Journal: Protein Sci. / Year: 1994Title: The NMR Solution Structure of the Pheromone Er-1 from the Ciliated Protozoan Euplotes Raikovi Authors: Mronga, S. / Luginbuhl, P. / Brown, L.R. / Ortenzi, C. / Luporini, P. / Bradshaw, R.A. / Wuthrich, K. #3:  Journal: J.Mol.Biol. / Year: 1990 Journal: J.Mol.Biol. / Year: 1990Title: Crystallization of the Euplotes Raikovi Mating Pheromone Er-1 Authors: Anderson, D. / Raffioni, S. / Luporini, P. / Bradshaw, R.A. / Eisenberg, D. #4:  Journal: J.Biol.Chem. / Year: 1988 Journal: J.Biol.Chem. / Year: 1988Title: Primary Structure of the Mating Pheromone Er-1 of the Ciliate Euplotes Raikovi Authors: Raffioni, S. / Luporini, P. / Chait, B.T. / Disper, S.S. / Bradshaw, R.A. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2erl.cif.gz 2erl.cif.gz | 38.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2erl.ent.gz pdb2erl.ent.gz | 27 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2erl.json.gz 2erl.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/er/2erl https://data.pdbj.org/pub/pdb/validation_reports/er/2erl ftp://data.pdbj.org/pub/pdb/validation_reports/er/2erl ftp://data.pdbj.org/pub/pdb/validation_reports/er/2erl | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 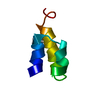
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein/peptide | Mass: 4417.881 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Euplotes raikovi (eukaryote) / References: UniProt: P10774 Euplotes raikovi (eukaryote) / References: UniProt: P10774 |
|---|---|
| #2: Chemical | ChemComp-EOH / |
| #3: Water | ChemComp-HOH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.53 Å3/Da / Density % sol: 19.6 % | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal | *PLUS Density % sol: 20 % | ||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 298-301 K / pH: 3.5 / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction source | Wavelength: 0.7107 |
|---|---|
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Sep 20, 1993 |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.7107 Å / Relative weight: 1 |
| Reflection | Resolution: 1→25.26 Å / Num. obs: 13659 / % possible obs: 93.8 % / Observed criterion σ(I): 0 / Redundancy: 7.5 % / Rmerge(I) obs: 0.054 |
| Reflection | *PLUS Lowest resolution: 8 Å / Num. obs: 13461 / % possible obs: 92.4 % / Num. measured all: 101188 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1→25.25 Å / Num. parameters: 3121 / Num. restraintsaints: 3883 / σ(F): 0 Stereochemistry target values: ENGH AND HUBER (1991) ACTA CRYST. A47, 392-400 Details: ANISOTROPIC TEMPERATURE FACTORS HAVE BEEN REFINED FOR THE NON-HYDROGEN ATOMS IN THIS ENTRY. HYDROGEN ATOMS HAVE ISOTROPIC U'S THAT ARE 1.2 TIMES THE ISOTROPIC U'S OF THE ATOMS ON WHICH THEY ...Details: ANISOTROPIC TEMPERATURE FACTORS HAVE BEEN REFINED FOR THE NON-HYDROGEN ATOMS IN THIS ENTRY. HYDROGEN ATOMS HAVE ISOTROPIC U'S THAT ARE 1.2 TIMES THE ISOTROPIC U'S OF THE ATOMS ON WHICH THEY RIDE. THE QUANTITIES ON THE ANISOU RECORD FOR EACH HYDROGEN ATOM ARE THE EQUIVALENT-TO-ISOTROPIC U'S.
| |||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: DRIESSEN, ET AL., (1989) J.APPL.CRYST. 22, 510-516 | |||||||||||||||||||||||||||||||||
| Refine analyze | Num. disordered residues: 4 / Occupancy sum non hydrogen: 1 | |||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1→25.25 Å
| |||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||
| Software | *PLUS Name: SHELXL / Version: 93 / Classification: refinement | |||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor Rwork: 0.129 / Lowest resolution: 26 Å / % reflection Rfree: 10 % / Rfactor obs: 0.1292 | |||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | |||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS Type: s_plane_restr / Dev ideal: 0.206 |
 Movie
Movie Controller
Controller











 PDBj
PDBj







