[English] 日本語
 Yorodumi
Yorodumi- PDB-2aza: STRUCTURE OF AZURIN FROM ALCALIGENES DENITRIFICANS. REFINEMENT AT... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2aza | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | STRUCTURE OF AZURIN FROM ALCALIGENES DENITRIFICANS. REFINEMENT AT 1.8 ANGSTROMS RESOLUTION AND COMPARISON OF THE TWO CRYSTALLOGRAPHICALLY INDEPENDENT MOLECULES | |||||||||
 Components Components | AZURIN | |||||||||
 Keywords Keywords | ELECTRON TRANSPORT PROTEIN(CUPROPROTEIN) | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Achromobacter xylosoxidans (bacteria) Achromobacter xylosoxidans (bacteria) | |||||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1.8 Å X-RAY DIFFRACTION / Resolution: 1.8 Å | |||||||||
 Authors Authors | Baker, E.N. / Norris, G.E. | |||||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1988 Journal: J.Mol.Biol. / Year: 1988Title: Structure of azurin from Alcaligenes denitrificans refinement at 1.8 A resolution and comparison of the two crystallographically independent molecules. Authors: Baker, E.N. #1:  Journal: J.Am.Chem.Soc. / Year: 1986 Journal: J.Am.Chem.Soc. / Year: 1986Title: Blue Copper Proteins. The Copper Site in Azurin from Alcaligenes Denitrificans Authors: Norris, G.E. / Anderson, B.F. / Baker, E.N. #2:  Journal: J.Mol.Biol. / Year: 1983 Journal: J.Mol.Biol. / Year: 1983Title: Structure of Azurin from Alcaligenes Denitrificans at 2.5 Angstroms Resolution Authors: Norris, G.E. / Anderson, B.F. / Baker, E.N. #3:  Journal: J.Mol.Biol. / Year: 1979 Journal: J.Mol.Biol. / Year: 1979Title: Purification and Preliminary Crystallographic Studies on Azurin and Cytochrome C(Prime) from Alcaligenes Denitrificans and Alcaligenes Sp. Ncib 11015 Authors: Norris, G.E. / Anderson, B.F. / Baker, E.N. / Rumball, S.V. | |||||||||
| History |
| |||||||||
| Remark 700 | SHEET STRAND 1 OF SHEET B1A (AND B1B) AND STRAND 1 OF SHEET B2A (AND B2B) ARE PARTS OF A PIECE OF ...SHEET STRAND 1 OF SHEET B1A (AND B1B) AND STRAND 1 OF SHEET B2A (AND B2B) ARE PARTS OF A PIECE OF EXTENDED CHAIN WHICH IS SPLIT BETWEEN BETWEEN THE TWO SHEETS. RESIDUES 14-16 BELONG TO SHEET B1 AND RESIDUES 18-22 BELONG TO SHEET B2 WITH A KINK IN BETWEEN. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2aza.cif.gz 2aza.cif.gz | 70.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2aza.ent.gz pdb2aza.ent.gz | 52 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2aza.json.gz 2aza.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/az/2aza https://data.pdbj.org/pub/pdb/validation_reports/az/2aza ftp://data.pdbj.org/pub/pdb/validation_reports/az/2aza ftp://data.pdbj.org/pub/pdb/validation_reports/az/2aza | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 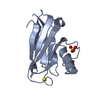
| |||||||||
| 2 | 
| |||||||||
| Unit cell |
| |||||||||
| Atom site foot note | 1: SEE REMARK 5. / 2: SEE REMARK 6. / 3: SEE REMARKS 5 AND 6. | |||||||||
| Components on special symmetry positions |
| |||||||||
| Noncrystallographic symmetry (NCS) | NCS oper: (Code: given Matrix: (-0.10158, 0.99439, 0.02952), Vector: Details | THE TRANSFORMATION PROVIDED ON THE *MTRIX* RECORDS BELOW WILL PRODUCE APPROXIMATE COORDINATES FOR MOLECULE 1 (CHAIN INDICATOR *A*) WHEN APPLIED TO THE COORDINATES OF MOLECULE 2 (CHAIN INDICATOR *B*). | |
- Components
Components
| #1: Protein | Mass: 13997.919 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Achromobacter xylosoxidans (bacteria) / References: UniProt: P00280 Achromobacter xylosoxidans (bacteria) / References: UniProt: P00280#2: Chemical | #3: Chemical | #4: Water | ChemComp-HOH / | Compound details | TURNS 3A AND 12A (AND 3B AND 12B) ARE MORE LIKE ALPHA-TURNS AS THEY HAVE GOOD 1-5 H-BONDS (O ALA 40 ...TURNS 3A AND 12A (AND 3B AND 12B) ARE MORE LIKE ALPHA-TURNS AS THEY HAVE GOOD 1-5 H-BONDS (O ALA 40 - N MET 44 AND O HIS 117 - N MET 121) WHICH ARE SHORTER THAN THE 1-4 H-BONDS (O ALA 40 - MET 43 AND 0 HIS 117 - N MET 120). | Has protein modification | Y | Sequence details | RESIDUE 42 IS INCLUDED HERE AS SER BOTH IN THE *ATOM* AND *SEQRES* RECORDS. RESIDUE 42 IS ALA IN ...RESIDUE 42 IS INCLUDED HERE AS SER BOTH IN THE *ATOM* AND *SEQRES* RECORDS. RESIDUE 42 IS ALA IN THE CHEMICALLY | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.47 Å3/Da / Density % sol: 50.24 % | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS pH: 5 / Method: other | |||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Reflection | *PLUS Highest resolution: 1.8 Å / Num. obs: 21980 / % possible obs: 87 % |
|---|
- Processing
Processing
| Software | Name: TNT / Classification: refinement | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Highest resolution: 1.8 Å Details: ONLY INTERNAL H-BONDS INVOLVING SIDE CHAINS OR CROSSLINKING H BONDS BETWEEN STRANDS ARE PRESENTED ON THE CONECT RECORDS BELOW.
| ||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 1.8 Å
| ||||||||||||
| Refine LS restraints |
| ||||||||||||
| Refinement | *PLUS Highest resolution: 1.8 Å / Lowest resolution: 10 Å / Rfactor all: 0.157 / Num. reflection all: 21980 | ||||||||||||
| Solvent computation | *PLUS | ||||||||||||
| Displacement parameters | *PLUS | ||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller











 PDBj
PDBj




