+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1uri | ||||||
|---|---|---|---|---|---|---|---|
| Title | AZURIN MUTANT WITH MET 121 REPLACED BY GLN | ||||||
 Components Components | AZURIN | ||||||
 Keywords Keywords | ELECTRON TRANSPORT / COPPER / PERIPLASMIC | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  Achromobacter denitrificans (bacteria) Achromobacter denitrificans (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1.94 Å X-RAY DIFFRACTION / Resolution: 1.94 Å | ||||||
 Authors Authors | Romero, A. / Nar, H. / Huber, R. / Messerschmidt, A. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1993 Journal: J.Mol.Biol. / Year: 1993Title: X-ray analysis and spectroscopic characterization of M121Q azurin. A copper site model for stellacyanin. Authors: Romero, A. / Hoitink, C.W. / Nar, H. / Huber, R. / Messerschmidt, A. / Canters, G.W. #1:  Journal: J.Mol.Biol. / Year: 1988 Journal: J.Mol.Biol. / Year: 1988Title: Structure of Azurin from Alcaligenes Denitrificans Authors: Baker, E.N. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1uri.cif.gz 1uri.cif.gz | 65.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1uri.ent.gz pdb1uri.ent.gz | 48.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1uri.json.gz 1uri.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ur/1uri https://data.pdbj.org/pub/pdb/validation_reports/ur/1uri ftp://data.pdbj.org/pub/pdb/validation_reports/ur/1uri ftp://data.pdbj.org/pub/pdb/validation_reports/ur/1uri | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 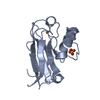
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 14004.936 Da / Num. of mol.: 2 / Mutation: M121Q Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Achromobacter denitrificans (bacteria) / Strain: JM101 / Plasmid: PCH5 / Production host: Achromobacter denitrificans (bacteria) / Strain: JM101 / Plasmid: PCH5 / Production host:  #2: Chemical | #3: Chemical | #4: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.46 Å3/Da / Density % sol: 49.97 % | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS pH: 5.2 / Method: vapor diffusion | |||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction source | Wavelength: 1.5418 |
|---|---|
| Detector | Type: ENRAF-NONIUS FAST / Detector: DIFFRACTOMETER / Date: Jun 2, 1991 |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Num. obs: 16824 / % possible obs: 79 % / Observed criterion σ(I): 2 / Rmerge(I) obs: 0.077 |
| Reflection | *PLUS Highest resolution: 1.94 Å / Lowest resolution: 9999 Å / Num. measured all: 58815 |
| Reflection shell | *PLUS Highest resolution: 1.94 Å / Lowest resolution: 1.99 Å / % possible obs: 27.2 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1.94→8 Å / σ(F): 2 Details: BESIDES GLYCINE RESIDUES, ONLY WEAKLY DEFINED RESIDUES FROM THE N-TERMINAL (GLN 2) LIE OUTSIDE THE ALLOWED REGIONS IN THE RAMACHANDRAN PLOT.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 21.4 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.94→8 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Classification: refinement X-PLOR / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Highest resolution: 1.94 Å / Lowest resolution: 1.99 Å / Rfactor all: 0.306 |
 Movie
Movie Controller
Controller












 PDBj
PDBj




