[English] 日本語
 Yorodumi
Yorodumi- PDB-2abk: REFINEMENT OF THE NATIVE STRUCTURE OF ENDONUCLEASE III TO A RESOL... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2abk | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | REFINEMENT OF THE NATIVE STRUCTURE OF ENDONUCLEASE III TO A RESOLUTION OF 1.85 ANGSTROM | |||||||||
 Components Components | ENDONUCLEASE III | |||||||||
 Keywords Keywords | ENDONUCLEASE / DNA-REPAIR / DNA GLYCOSYLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationoxidized pyrimidine nucleobase lesion DNA N-glycosylase activity / base-excision repair, AP site formation via deaminated base removal / base-excision repair, AP site formation / DNA N-glycosylase activity / uracil DNA N-glycosylase activity / DNA-(apurinic or apyrimidinic site) endonuclease activity / class I DNA-(apurinic or apyrimidinic site) endonuclease activity / DNA-(apurinic or apyrimidinic site) lyase / cellular response to UV / 4 iron, 4 sulfur cluster binding ...oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity / base-excision repair, AP site formation via deaminated base removal / base-excision repair, AP site formation / DNA N-glycosylase activity / uracil DNA N-glycosylase activity / DNA-(apurinic or apyrimidinic site) endonuclease activity / class I DNA-(apurinic or apyrimidinic site) endonuclease activity / DNA-(apurinic or apyrimidinic site) lyase / cellular response to UV / 4 iron, 4 sulfur cluster binding / DNA binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1.85 Å X-RAY DIFFRACTION / Resolution: 1.85 Å | |||||||||
 Authors Authors | Thayer, M.M. / Tainer, J.A. | |||||||||
 Citation Citation |  Journal: EMBO J. / Year: 1995 Journal: EMBO J. / Year: 1995Title: Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. Authors: Thayer, M.M. / Ahern, H. / Xing, D. / Cunningham, R.P. / Tainer, J.A. #1:  Journal: J.Mol.Biol. / Year: 1992 Journal: J.Mol.Biol. / Year: 1992Title: Crystallization and Crystallographic Characterization of the Iron-Sulfur Containing DNA-Repair Enzyme Endonuclease III from Escherichia Coli Enzyme Endonuclease III Authors: Kuo, C.-F. / Mcree, D.E. / Cunningham, R.P. / Tainer, J.A. #2:  Journal: Science / Year: 1992 Journal: Science / Year: 1992Title: Atomic Structure of the DNA Repair [4Fe-4S] Enzyme Endonuclease III Authors: Kuo, C.-F. / Mcree, D.E. / Fisher, C.L. / Handley, S. / Cunningham, R.P. / Tainer, J.A. | |||||||||
| History |
| |||||||||
| Remark 650 | HELIX DETERMINATION METHOD: KABSCH AND SANDER AS IMPLEMENTED BY PROCHECK FOLLOWED BY VISUAL ...HELIX DETERMINATION METHOD: KABSCH AND SANDER AS IMPLEMENTED BY PROCHECK FOLLOWED BY VISUAL INSPECTION OF MODEL. TURN KABSCH AND SANDER IN PROCHECK TURN_ID: FG, HAIRPIN OF HELIX HAIRPIN HELIX MOTIF. |
- Structure visualization
Structure visualization
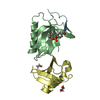
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2abk.cif.gz 2abk.cif.gz | 55.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2abk.ent.gz pdb2abk.ent.gz | 40.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2abk.json.gz 2abk.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ab/2abk https://data.pdbj.org/pub/pdb/validation_reports/ab/2abk ftp://data.pdbj.org/pub/pdb/validation_reports/ab/2abk ftp://data.pdbj.org/pub/pdb/validation_reports/ab/2abk | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 23595.398 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  References: UniProt: P0AB83, DNA-(apurinic or apyrimidinic site) lyase |
|---|---|
| #2: Chemical | ChemComp-SF4 / |
| #3: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.93 Å3/Da / Density % sol: 58.07 % | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS pH: 7 / Method: unknown | ||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction source | Wavelength: 1.5418 Å |
|---|---|
| Detector | Type: XENTRONICS / Detector: AREA DETECTOR / Date: May 18, 1993 |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.85→20 Å / Num. obs: 27787 / % possible obs: 99.8 % / Observed criterion σ(I): 0 / Redundancy: 10 % / Rmerge(I) obs: 0.07 |
| Reflection | *PLUS Rmerge(I) obs: 0.07 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1.85→10 Å / σ(F): 2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 23.7 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.85→10 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3 / Classification: refinement X-PLOR / Version: 3 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller











 PDBj
PDBj



