[English] 日本語
 Yorodumi
Yorodumi- PDB-1zwt: Structure of the globular head domain of the bundlin, BfpA, of th... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1zwt | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of the globular head domain of the bundlin, BfpA, of the bundle-forming pilus of Enteropathogenic E.coli | ||||||
 Components Components | Major structural subunit of bundle-forming pilus | ||||||
 Keywords Keywords | CELL ADHESION / alpha-beta fold / beta-sandwich / one disulfide bond | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  | ||||||
| Method | SOLUTION NMR / Simulated annealing with NMR restraints using ARIA | ||||||
 Authors Authors | Ramboarina, S. / Fernandes, P.J. / Daniell, S. / Islam, S. / Frankel, G. / Booy, F. / Donnenberg, M.S. / Matthews, S. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2005 Journal: J.Biol.Chem. / Year: 2005Title: Structure of the Bundle-forming Pilus from Enteropathogenic Escherichia coli Authors: Ramboarina, S. / Fernandes, P.J. / Daniell, S. / Islam, S. / Simpson, P. / Frankel, G. / Booy, F. / Donnenberg, M.S. / Matthews, S. #1: Journal: J.BIOMOL.NMR / Year: 2004 Title: Complete resonance assignments of bundlin (BfpA) from the bundle-forming pilus of enteropathogenic Escherichia coli Authors: Ramboarina, S. / Fernandes, P. / Simpson, P. / Frankel, G. / Donnenberg, M. / Matthews, S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1zwt.cif.gz 1zwt.cif.gz | 591 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1zwt.ent.gz pdb1zwt.ent.gz | 498.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1zwt.json.gz 1zwt.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zw/1zwt https://data.pdbj.org/pub/pdb/validation_reports/zw/1zwt ftp://data.pdbj.org/pub/pdb/validation_reports/zw/1zwt ftp://data.pdbj.org/pub/pdb/validation_reports/zw/1zwt | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 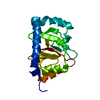
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 16486.092 Da / Num. of mol.: 1 / Fragment: BfpA Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||
| NMR details | Text: Hydrogen-exchange experiments |
- Sample preparation
Sample preparation
| Details | Contents: 0.5 mM BfpA U-15N,13C, 20 mM sodium phosphate buffer pH 5.2, 90%H20, 10% D2O Solvent system: 90%H20, 10% D2O |
|---|---|
| Sample conditions | Ionic strength: no salt / pH: 5.2 / Pressure: normal / Temperature: 303 K |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Radiation wavelength | Relative weight: 1 | |||||||||||||||
| NMR spectrometer |
|
- Processing
Processing
| NMR software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: Simulated annealing with NMR restraints using ARIA / Software ordinal: 1 Details: 577 Ambiguous and 2277 unambiguous NOE restraints, 86 TALOS constraints and 38 hydrogen bonds were imposed during ARIA structure calculations. | ||||||||||||||||
| NMR representative | Selection criteria: least number of violations, lowest energy | ||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with acceptable covalent geometry, structures with the least restraint violations, structures with the lowest energy Conformers calculated total number: 100 / Conformers submitted total number: 12 |
 Movie
Movie Controller
Controller












 PDBj
PDBj
 NMRPipe
NMRPipe