[English] 日本語
 Yorodumi
Yorodumi- PDB-1zbn: Solution structure of BIV TAR hairpin complexed to JDV Tat argini... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1zbn | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution structure of BIV TAR hairpin complexed to JDV Tat arginine-rich motif | ||||||
 Components Components |
| ||||||
 Keywords Keywords | RNA binding protein/RNA / RNA-peptide complex / RNA binding protein-RNA COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of viral transcription / host cell nucleolus / RNA-binding transcription regulator activity / RNA binding Similarity search - Function | ||||||
| Method | SOLUTION NMR / distance geometry, restrained molecular dynamics | ||||||
 Authors Authors | Calabro, V. / Daugherty, M.D. / Frankel, A.D. | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.Usa / Year: 2005 Journal: Proc.Natl.Acad.Sci.Usa / Year: 2005Title: A single intermolecular contact mediates intramolecular stabilization of both RNA and protein. Authors: Calabro, V. / Daugherty, M.D. / Frankel, A.D. #1:  Journal: Mol.Cell / Year: 2000 Journal: Mol.Cell / Year: 2000Title: An RNA-binding chameleon Authors: Smith, C.A. / Calabro, V. / Frankel, A.D. #2:  Journal: Science / Year: 1995 Journal: Science / Year: 1995Title: Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex Authors: Puglisi, J.D. / Chen, L. / Blanchard, S. / Frankel, A.D. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1zbn.cif.gz 1zbn.cif.gz | 243.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1zbn.ent.gz pdb1zbn.ent.gz | 201.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1zbn.json.gz 1zbn.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1zbn_validation.pdf.gz 1zbn_validation.pdf.gz | 364.8 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1zbn_full_validation.pdf.gz 1zbn_full_validation.pdf.gz | 504.3 KB | Display | |
| Data in XML |  1zbn_validation.xml.gz 1zbn_validation.xml.gz | 19.6 KB | Display | |
| Data in CIF |  1zbn_validation.cif.gz 1zbn_validation.cif.gz | 30.5 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zb/1zbn https://data.pdbj.org/pub/pdb/validation_reports/zb/1zbn ftp://data.pdbj.org/pub/pdb/validation_reports/zb/1zbn ftp://data.pdbj.org/pub/pdb/validation_reports/zb/1zbn | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 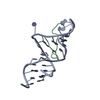
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
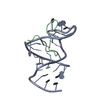
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 8923.310 Da / Num. of mol.: 1 / Mutation: A4G, U31C / Source method: obtained synthetically Details: This sequence occurs naturally in Bovine immunodeficiency virus (BIV). |
|---|---|
| #2: Protein/peptide | Mass: 2064.493 Da / Num. of mol.: 1 / Fragment: Arginine-rich domain / Source method: obtained synthetically Details: This sequence occurs naturally in Jembrana disease virus (JDV). References: UniProt: Q82854*PLUS |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||||||
| NMR details | Text: This structure was determined using standard 2D homonuclear techniques. |
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
|
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Varian UNITY / Manufacturer: Varian / Model: UNITY / Field strength: 600 MHz |
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: distance geometry, restrained molecular dynamics / Software ordinal: 1 Details: Structures were calculated beginning with 1,000 random structures and incrementally adding distance constraints in four iterations. The final 30 structures with the lowest number of ...Details: Structures were calculated beginning with 1,000 random structures and incrementally adding distance constraints in four iterations. The final 30 structures with the lowest number of violations were subjected to RMD. The structures are based on a total of 700 distance constraints, 61 dihedral angle restraints. | ||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 30 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller












 PDBj
PDBj





























 CYANA
CYANA