[English] 日本語
 Yorodumi
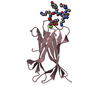
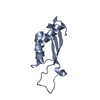
Yorodumi- PDB-1mnb: BIV TAT PEPTIDE (RESIDUES 68-81), NMR, MINIMIZED AVERAGE STRUCTURE -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1mnb | ||||||
|---|---|---|---|---|---|---|---|
| Title | BIV TAT PEPTIDE (RESIDUES 68-81), NMR, MINIMIZED AVERAGE STRUCTURE | ||||||
 Components Components |
| ||||||
 Keywords Keywords | Viral protein/RNA / COMPLEX (REGULATORY PROTEIN-RNA) / TRANSCRIPTION REGULATION / Viral protein-RNA COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of viral transcription / host cell nucleolus / RNA-binding transcription regulator activity / RNA binding Similarity search - Function | ||||||
| Method | SOLUTION NMR | ||||||
 Authors Authors | Puglisi, J.D. / Chen, L. / Blanchard, S. / Frankel, A.D. | ||||||
 Citation Citation |  Journal: Science / Year: 1995 Journal: Science / Year: 1995Title: Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex. Authors: Puglisi, J.D. / Chen, L. / Blanchard, S. / Frankel, A.D. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1mnb.cif.gz 1mnb.cif.gz | 37 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1mnb.ent.gz pdb1mnb.ent.gz | 22.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1mnb.json.gz 1mnb.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mn/1mnb https://data.pdbj.org/pub/pdb/validation_reports/mn/1mnb ftp://data.pdbj.org/pub/pdb/validation_reports/mn/1mnb ftp://data.pdbj.org/pub/pdb/validation_reports/mn/1mnb | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 8923.310 Da / Num. of mol.: 1 / Fragment: RESIDUES 4 - 32 / Source method: obtained synthetically |
|---|---|
| #2: Protein/peptide | Mass: 1730.083 Da / Num. of mol.: 1 / Fragment: RESIDUES 68 - 81 Source method: isolated from a genetically manipulated source References: UniProt: P19564 |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|---|
| NMR details | Text: 50 MM NACL 10MM NA PHOSPHATE |
- Sample preparation
Sample preparation
| Sample conditions | pH: 6.4 / Temperature: 298 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
- Processing
Processing
| NMR software | Name: NMRARCHITECT, DISCOVER / Version: DISCOVER / Developer: BIOSYM / Classification: refinement |
|---|---|
| NMR ensemble | Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller










 PDBj
PDBj





























