[English] 日本語
 Yorodumi
Yorodumi- PDB-1vg0: The crystal structures of the REP-1 protein in complex with monop... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1vg0 | ||||||
|---|---|---|---|---|---|---|---|
| Title | The crystal structures of the REP-1 protein in complex with monoprenylated Rab7 protein | ||||||
 Components Components |
| ||||||
 Keywords Keywords | PROTEIN BINDING/PROTEIN TRANSPORT / RAB PRENYLATION / POST-TRANSLATIONAL MODIFICATION / PROTEIN BINDING-PROTEIN TRANSPORT COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationRAB GEFs exchange GTP for GDP on RABs / RHOF GTPase cycle / TBC/RABGAPs / : / synaptic vesicle recycling via endosome / RAC2 GTPase cycle / RHOH GTPase cycle / phagosome acidification / TP53 regulates transcription of several additional cell death genes whose specific roles in p53-dependent apoptosis remain uncertain / RHOG GTPase cycle ...RAB GEFs exchange GTP for GDP on RABs / RHOF GTPase cycle / TBC/RABGAPs / : / synaptic vesicle recycling via endosome / RAC2 GTPase cycle / RHOH GTPase cycle / phagosome acidification / TP53 regulates transcription of several additional cell death genes whose specific roles in p53-dependent apoptosis remain uncertain / RHOG GTPase cycle / lipophagy / protein to membrane docking / positive regulation of viral process / RAB geranylgeranylation / Rab-protein geranylgeranyltransferase complex / RHOQ GTPase cycle / neurotransmitter receptor transport, postsynaptic endosome to lysosome / epidermal growth factor catabolic process / alveolar lamellar body / RAC1 GTPase cycle / phagosome-lysosome fusion / negative regulation of exosomal secretion / establishment of vesicle localization / protein geranylgeranylation / MHC class II antigen presentation / retromer complex binding / presynaptic endosome / vesicle-mediated transport in synapse / retromer complex / exocytic vesicle / protein localization to lysosome / endosome to plasma membrane protein transport / early endosome to late endosome transport / phagophore assembly site membrane / positive regulation of exosomal secretion / melanosome membrane / protein targeting to lysosome / retrograde transport, endosome to Golgi / protein targeting to membrane / Neutrophil degranulation / GDP-dissociation inhibitor activity / small GTPase-mediated signal transduction / endosome to lysosome transport / blood vessel development / autophagosome membrane / viral release from host cell / autophagosome assembly / bone resorption / lipid catabolic process / vesicle-mediated transport / phagocytic vesicle / lipid droplet / GTPase activator activity / small monomeric GTPase / response to bacterium / intracellular protein transport / small GTPase binding / mitochondrial membrane / phagocytic vesicle membrane / terminal bouton / positive regulation of protein catabolic process / GDP binding / late endosome / synaptic vesicle membrane / late endosome membrane / G protein activity / lysosome / endosome / endosome membrane / lysosomal membrane / GTPase activity / GTP binding / protein-containing complex binding / glutamatergic synapse / Golgi apparatus / mitochondrion / metal ion binding / nucleus / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å MOLECULAR REPLACEMENT / Resolution: 2.2 Å | ||||||
 Authors Authors | Rak, A. / Pylypenko, O. / Niculae, A. / Pyatkov, K. / Goody, R.S. / Alexandrov, K. | ||||||
 Citation Citation |  Journal: Cell(Cambridge,Mass.) / Year: 2004 Journal: Cell(Cambridge,Mass.) / Year: 2004Title: Structure of the Rab7:REP-1 complex: insights into the mechanism of Rab prenylation and choroideremia disease Authors: Rak, A. / Pylypenko, O. / Niculae, A. / Pyatkov, K. / Goody, R.S. / Alexandrov, K. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1vg0.cif.gz 1vg0.cif.gz | 162.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1vg0.ent.gz pdb1vg0.ent.gz | 121.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1vg0.json.gz 1vg0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vg/1vg0 https://data.pdbj.org/pub/pdb/validation_reports/vg/1vg0 ftp://data.pdbj.org/pub/pdb/validation_reports/vg/1vg0 ftp://data.pdbj.org/pub/pdb/validation_reports/vg/1vg0 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1vg1C  1vg8C  1vg9C  1ltxS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 2 types, 2 molecules AB
| #1: Protein | Mass: 72622.234 Da / Num. of mol.: 1 / Mutation: K231Q, K462R, T473A, A483G Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #2: Protein | Mass: 23530.744 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
-Non-polymers , 6 types, 443 molecules 
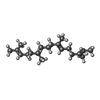









| #3: Chemical | ChemComp-CL / |
|---|---|
| #4: Chemical | ChemComp-GER / |
| #5: Chemical | ChemComp-MG / |
| #6: Chemical | ChemComp-GDP / |
| #7: Chemical | ChemComp-PG4 / |
| #8: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.47 Å3/Da / Density % sol: 50.2 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 7.2 Details: 23% PEG 3350, 0.4M diAmmonium Tartrate, pH 7.2, VAPOR DIFFUSION, HANGING DROP, temperature 298K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-4 / Wavelength: 0.9393 Å / Beamline: ID14-4 / Wavelength: 0.9393 Å |
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Feb 28, 2002 |
| Radiation | Monochromator: Si 111 CHANNEL / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9393 Å / Relative weight: 1 |
| Reflection | Resolution: 2.2→18 Å / Num. all: 45399 / Num. obs: 45399 / % possible obs: 97.6 % / Observed criterion σ(F): 1 / Observed criterion σ(I): 1 / Redundancy: 3.6 % / Biso Wilson estimate: 22 Å2 / Rsym value: 0.081 / Net I/σ(I): 10.19 |
| Reflection shell | Resolution: 2.2→2.3 Å / Redundancy: 3.6 % / Mean I/σ(I) obs: 3.61 / Num. unique all: 5630 / Rsym value: 0.357 / % possible all: 98.5 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: REP-1 from RabGGTase:REP-1 complex (PDB code 1LTX) Resolution: 2.2→18 Å / Rfactor Rfree error: 0.005 / Data cutoff high absF: 2781170.14 / Data cutoff low absF: 0 / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 33.7874 Å2 / ksol: 0.325713 e/Å3 | ||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.2→18 Å
| ||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.2→2.34 Å / Rfactor Rfree error: 0.015 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj














