[English] 日本語
 Yorodumi
Yorodumi- PDB-1ltx: Structure of Rab Escort Protein-1 in complex with Rab geranylgera... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ltx | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of Rab Escort Protein-1 in complex with Rab geranylgeranyl transferase and isoprenoid | ||||||
 Components Components |
| ||||||
 Keywords Keywords | Transferase/Protein binding / RAB PRENYLATION / PRENYLTRANSFERASE / LUCINE-RICH REPEATS / POST-TRANSLATIONAL MODIFICATION / Transferase-Protein binding COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationRAB GEFs exchange GTP for GDP on RABs / TP53 regulates transcription of several additional cell death genes whose specific roles in p53-dependent apoptosis remain uncertain / protein geranylgeranyltransferase type II / RAB geranylgeranylation / Rab-protein geranylgeranyltransferase complex / isoprenoid binding / Rab geranylgeranyltransferase activity / protein geranylgeranylation / protein targeting to membrane / GDP-dissociation inhibitor activity ...RAB GEFs exchange GTP for GDP on RABs / TP53 regulates transcription of several additional cell death genes whose specific roles in p53-dependent apoptosis remain uncertain / protein geranylgeranyltransferase type II / RAB geranylgeranylation / Rab-protein geranylgeranyltransferase complex / isoprenoid binding / Rab geranylgeranyltransferase activity / protein geranylgeranylation / protein targeting to membrane / GDP-dissociation inhibitor activity / small GTPase-mediated signal transduction / blood vessel development / endoplasmic reticulum to Golgi vesicle-mediated transport / vesicle-mediated transport / GTPase activator activity / intracellular protein transport / small GTPase binding / protein-containing complex binding / zinc ion binding / nucleus / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.7 Å MOLECULAR REPLACEMENT / Resolution: 2.7 Å | ||||||
 Authors Authors | Pylypenko, O. / Rak, A. / Reents, R. / Niculae, A. / Thoma, N.H. / Waldmann, H. / Schlichting, I. / Goody, R.S. / Alexandrov, K. | ||||||
 Citation Citation |  Journal: Mol.Cell / Year: 2003 Journal: Mol.Cell / Year: 2003Title: Structure of Rab Escort Protein-1 in Complex with Rab Geranylgeranyltransferase Authors: Pylypenko, O. / Rak, A. / Reents, R. / Niculae, A. / Sidorovitch, V. / Cioaca, M.D. / Bessolitsyna, E. / Thoma, N.H. / Waldmann, H. / Schlichting, I. / Goody, R.S. / Alexandrov, K. #1:  Journal: J.STRUCT.BIOL. / Year: 2001 Journal: J.STRUCT.BIOL. / Year: 2001Title: Crystallization and Preliminary X-ray Diffraction Analysis of the Rab Escort Protein-1 in Complex with Rab Geranylgeranyltransferase Authors: Rak, A. / Reents, R. / Pylypenko, O. / Niculae, A. / Sidorovitch, V. / Thoma, N.H. / Waldmann, H. / Schlichting, I. / Goody, R.S. / Alexandrov, K. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ltx.cif.gz 1ltx.cif.gz | 281.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ltx.ent.gz pdb1ltx.ent.gz | 222.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ltx.json.gz 1ltx.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/lt/1ltx https://data.pdbj.org/pub/pdb/validation_reports/lt/1ltx ftp://data.pdbj.org/pub/pdb/validation_reports/lt/1ltx ftp://data.pdbj.org/pub/pdb/validation_reports/lt/1ltx | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1dceS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-RAB GERANYLGERANYLTRANSFERASE ... , 2 types, 2 molecules AB
| #1: Protein | Mass: 64981.156 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q08602, Transferases; Transferring alkyl or aryl groups, other than methyl groups |
|---|---|
| #2: Protein | Mass: 36892.160 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q08603, Transferases; Transferring alkyl or aryl groups, other than methyl groups |
-Protein / Protein/peptide , 2 types, 2 molecules RP
| #3: Protein | Mass: 72594.219 Da / Num. of mol.: 1 / Mutation: A473T, G483A, Q231K Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #4: Protein/peptide | Mass: 302.326 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: PUTATIVE PEPTIDE. |
-Non-polymers , 4 types, 106 molecules 

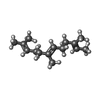




| #5: Chemical | ChemComp-ZN / |
|---|---|
| #6: Chemical | ChemComp-CL / |
| #7: Chemical | ChemComp-FAR / |
| #8: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.05 Å3/Da / Density % sol: 59.65 % | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 7.2 Details: 20% PEG 3350, 100mM KSCN, 100mM Namalonate, pH 7.2, VAPOR DIFFUSION, HANGING DROP, temperature 298.0K | |||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusionDetails: used microseeding, Rak, A., (2001) J.STRUCT.BIOL., 136, 158. | |||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-1 / Wavelength: 0.9393 Å / Beamline: ID14-1 / Wavelength: 0.9393 Å |
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9393 Å / Relative weight: 1 |
| Reflection | Resolution: 2.7→19.7 Å / Num. all: 54362 / Num. obs: 54362 / % possible obs: 94.5 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 3.6 % / Biso Wilson estimate: 53.8 Å2 / Rsym value: 0.094 / Net I/σ(I): 9.5 |
| Reflection shell | Resolution: 2.7→2.8 Å / Redundancy: 2.7 % / Mean I/σ(I) obs: 3.25 / Num. unique all: 3716 / Rsym value: 0.364 / % possible all: 62.5 |
| Reflection | *PLUS Rmerge(I) obs: 0.094 |
| Reflection shell | *PLUS % possible obs: 62.5 % / Num. unique obs: 3716 / Rmerge(I) obs: 0.364 / Mean I/σ(I) obs: 3.2 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1DCE Resolution: 2.7→19.74 Å / Rfactor Rfree error: 0.005 / Data cutoff high absF: 2824477.81 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 31.0783 Å2 / ksol: 0.342551 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 52.4 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.7→19.74 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.7→2.87 Å / Rfactor Rfree error: 0.022 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2.8 Å / % reflection Rfree: 5 % / Rfactor Rfree: 0.274 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Highest resolution: 2.7 Å / Lowest resolution: 2.8 Å |
 Movie
Movie Controller
Controller










 PDBj
PDBj




