[English] 日本語
 Yorodumi
Yorodumi- PDB-1tl7: Complex Of Gs- With The Catalytic Domains Of Mammalian Adenylyl C... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1tl7 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Complex Of Gs- With The Catalytic Domains Of Mammalian Adenylyl Cyclase: Complex With 2'(3')-O-(N-methylanthraniloyl)-guanosine 5'-triphosphate and Mn | ||||||
 Components Components |
| ||||||
 Keywords Keywords | LYASE / adenylyl cyclase / Gsa / MANT-GTP | ||||||
| Function / homology |  Function and homology information Function and homology informationAdenylate cyclase activating pathway / Hedgehog 'off' state / PKA activation / Adenylate cyclase inhibitory pathway / sensory perception of chemical stimulus / adenylate cyclase / regulation of insulin secretion involved in cellular response to glucose stimulus / mu-type opioid receptor binding / corticotropin-releasing hormone receptor 1 binding / cAMP biosynthetic process ...Adenylate cyclase activating pathway / Hedgehog 'off' state / PKA activation / Adenylate cyclase inhibitory pathway / sensory perception of chemical stimulus / adenylate cyclase / regulation of insulin secretion involved in cellular response to glucose stimulus / mu-type opioid receptor binding / corticotropin-releasing hormone receptor 1 binding / cAMP biosynthetic process / adenylate cyclase activity / G alpha (z) signalling events / beta-2 adrenergic receptor binding / adenylate cyclase binding / D1 dopamine receptor binding / adenylate cyclase-activating adrenergic receptor signaling pathway / insulin-like growth factor receptor binding / ionotropic glutamate receptor binding / cellular response to forskolin / adenylate cyclase activator activity / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein beta/gamma-subunit complex binding / adenylate cyclase-activating G protein-coupled receptor signaling pathway / adenylate cyclase-activating dopamine receptor signaling pathway / heterotrimeric G-protein complex / manganese ion binding / positive regulation of cytosolic calcium ion concentration / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / intracellular signal transduction / cilium / membrane raft / GTPase activity / dendrite / GTP binding / magnesium ion binding / protein-containing complex / ATP binding / metal ion binding / membrane / plasma membrane / cytoplasm Similarity search - Function | ||||||
| Biological species |    | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.8 Å MOLECULAR REPLACEMENT / Resolution: 2.8 Å | ||||||
 Authors Authors | Mou, T.C. / Gille, A. / Seifert, R.J. / Sprang, S.R. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2005 Journal: J.Biol.Chem. / Year: 2005Title: Structural basis for the inhibition of mammalian membrane adenylyl cyclase by 2 '(3')-O-(N-Methylanthraniloyl)-guanosine 5 '-triphosphate. Authors: Mou, T.C. / Gille, A. / Fancy, D.A. / Seifert, R. / Sprang, S.R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1tl7.cif.gz 1tl7.cif.gz | 164.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1tl7.ent.gz pdb1tl7.ent.gz | 123.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1tl7.json.gz 1tl7.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1tl7_validation.pdf.gz 1tl7_validation.pdf.gz | 1.4 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1tl7_full_validation.pdf.gz 1tl7_full_validation.pdf.gz | 1.5 MB | Display | |
| Data in XML |  1tl7_validation.xml.gz 1tl7_validation.xml.gz | 30.4 KB | Display | |
| Data in CIF |  1tl7_validation.cif.gz 1tl7_validation.cif.gz | 40.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/tl/1tl7 https://data.pdbj.org/pub/pdb/validation_reports/tl/1tl7 ftp://data.pdbj.org/pub/pdb/validation_reports/tl/1tl7 ftp://data.pdbj.org/pub/pdb/validation_reports/tl/1tl7 | HTTPS FTP |
-Related structure data
| Related structure data |  1u0hC  1azsS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Adenylate cyclase, type ... , 2 types, 2 molecules AB
| #1: Protein | Mass: 24495.361 Da / Num. of mol.: 1 / Fragment: C1A Domain Of Adenylyl Cyclase Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #2: Protein | Mass: 23717.033 Da / Num. of mol.: 1 / Fragment: C2A Domain Of Adenylyl Cyclase Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
-Protein , 1 types, 1 molecules C
| #3: Protein | Mass: 46656.438 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|
-Non-polymers , 7 types, 25 molecules 

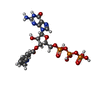










| #4: Chemical | | #5: Chemical | ChemComp-FOK / | #6: Chemical | ChemComp-ONM / | #7: Chemical | ChemComp-MG / | #8: Chemical | ChemComp-CL / | #9: Chemical | ChemComp-GSP / | #10: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.1 Å3/Da / Density % sol: 60.1 % |
|---|---|
| Crystal grow | Temperature: 289 K / Method: vapor diffusion, hanging drop / pH: 5.6 Details: 7.5-7.8% PEG 8000, 0.5M NACL, 0.1M PHOSPHATE BUFFER, pH 5.6, VAPOR DIFFUSION, HANGING DROP, temperature 289K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-BM / Wavelength: 1.0393 Å / Beamline: 19-BM / Wavelength: 1.0393 Å |
| Detector | Type: SBC-3 / Detector: CCD / Date: Nov 23, 2003 Details: Active area: 210 x 210 mm2 Pixel sizes: 0.079 mm (unbinned mode) and 0.159 mm (2 x 2 binned mode) Unbinned images: 3072 pixels x 3072 pixels 2 x 2 Binned images: 1536 pixels x 1536 pixels |
| Radiation | Monochromator: Si 111 / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.0393 Å / Relative weight: 1 |
| Reflection | Resolution: 2.8→50 Å / Num. all: 25538 / Num. obs: 22694 / Observed criterion σ(F): -2 / Observed criterion σ(I): -2 / Redundancy: 3.4 % / Rsym value: 0.162 / Net I/σ(I): 7.34 |
| Reflection shell | Resolution: 2.8→2.91 Å / Redundancy: 1.7 % / Rmerge(I) obs: 0.379 / Mean I/σ(I) obs: 1.44 / Rsym value: 0.378 / % possible all: 66.2 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PBD entry 1AZS Resolution: 2.8→14.99 Å / Rfactor Rfree error: 0.009 / Data cutoff high absF: 167322.76 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 10 Å2 / ksol: 0.267686 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 45.2 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.8→14.99 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.8→2.97 Å / Rfactor Rfree error: 0.035 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller


















 PDBj
PDBj














