[English] 日本語
 Yorodumi
Yorodumi- PDB-1qe6: INTERLEUKIN-8 WITH AN ADDED DISULFIDE BETWEEN RESIDUES 5 AND 33 (... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1qe6 | ||||||
|---|---|---|---|---|---|---|---|
| Title | INTERLEUKIN-8 WITH AN ADDED DISULFIDE BETWEEN RESIDUES 5 AND 33 (L5C/H33C) | ||||||
 Components Components | INTERLEUKIN-8 VARIANT | ||||||
 Keywords Keywords | IMMUNE SYSTEM / INTERCRINE ALPHA FAMILY | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of single stranded viral RNA replication via double stranded DNA intermediate / regulation of entry of bacterium into host cell / interleukin-8 receptor binding / negative regulation of cell adhesion molecule production / CXCR chemokine receptor binding / ATF4 activates genes in response to endoplasmic reticulum stress / embryonic digestive tract development / neutrophil activation / induction of positive chemotaxis / positive regulation of neutrophil chemotaxis ...regulation of single stranded viral RNA replication via double stranded DNA intermediate / regulation of entry of bacterium into host cell / interleukin-8 receptor binding / negative regulation of cell adhesion molecule production / CXCR chemokine receptor binding / ATF4 activates genes in response to endoplasmic reticulum stress / embryonic digestive tract development / neutrophil activation / induction of positive chemotaxis / positive regulation of neutrophil chemotaxis / chemokine activity / Chemokine receptors bind chemokines / negative regulation of G protein-coupled receptor signaling pathway / Interleukin-10 signaling / cellular response to interleukin-1 / regulation of cell adhesion / cellular response to fibroblast growth factor stimulus / neutrophil chemotaxis / Peptide ligand-binding receptors / response to endoplasmic reticulum stress / calcium-mediated signaling / response to molecule of bacterial origin / receptor internalization / positive regulation of angiogenesis / chemotaxis / antimicrobial humoral immune response mediated by antimicrobial peptide / cellular response to tumor necrosis factor / heparin binding / cellular response to lipopolysaccharide / Senescence-Associated Secretory Phenotype (SASP) / angiogenesis / Interleukin-4 and Interleukin-13 signaling / G alpha (i) signalling events / intracellular signal transduction / G protein-coupled receptor signaling pathway / inflammatory response / negative regulation of cell population proliferation / negative regulation of gene expression / positive regulation of gene expression / signal transduction / extracellular space / extracellular region Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2.35 Å SYNCHROTRON / Resolution: 2.35 Å | ||||||
 Authors Authors | Gerber, N. / Lowman, H. / Artis, D.R. / Eigenbrot, C. | ||||||
 Citation Citation |  Journal: Proteins / Year: 2000 Journal: Proteins / Year: 2000Title: Receptor-binding conformation of the "ELR" motif of IL-8: X-ray structure of the L5C/H33C variant at 2.35 A resolution. Authors: Gerber, N. / Lowman, H. / Artis, D.R. / Eigenbrot, C. #1:  Journal: Proc.Natl.Acad.Sci.USA / Year: 1991 Journal: Proc.Natl.Acad.Sci.USA / Year: 1991Title: Crystal structure of IL-8:Symbiosis of NMR and crystallography Authors: Baldwin, E.T. / Weber, I.T. / Charles, R.St. / Xuan, J.-C. #2:  Journal: Proteins / Year: 1997 Journal: Proteins / Year: 1997Title: Structural change and receptor binding in a chemokine mutant with a re- arranged disulfide: X-ray structure of E38C/C50A IL-8 at 2 A resolution. Authors: Eigenbrot, C. / Lowman, H.B. / Chee, L. / Artis, D.R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1qe6.cif.gz 1qe6.cif.gz | 71.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1qe6.ent.gz pdb1qe6.ent.gz | 54 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1qe6.json.gz 1qe6.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1qe6_validation.pdf.gz 1qe6_validation.pdf.gz | 391.4 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1qe6_full_validation.pdf.gz 1qe6_full_validation.pdf.gz | 395.3 KB | Display | |
| Data in XML |  1qe6_validation.xml.gz 1qe6_validation.xml.gz | 6.9 KB | Display | |
| Data in CIF |  1qe6_validation.cif.gz 1qe6_validation.cif.gz | 11.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qe/1qe6 https://data.pdbj.org/pub/pdb/validation_reports/qe/1qe6 ftp://data.pdbj.org/pub/pdb/validation_reports/qe/1qe6 ftp://data.pdbj.org/pub/pdb/validation_reports/qe/1qe6 | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 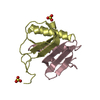
| ||||||||
| 3 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 8356.787 Da / Num. of mol.: 4 / Mutation: L5C, H33C Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Cell: MONOCYTE / Production host: Homo sapiens (human) / Cell: MONOCYTE / Production host:  #2: Chemical | #3: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.33 Å3/Da / Density % sol: 47.22 % | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 292 K / Method: vapor diffusion, hanging drop / pH: 6.2 Details: NACL, AMMONIUM SULFATE, PEG 8000, pH 6.2, VAPOR DIFFUSION, HANGING DROP, temperature 19K | ||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 20 ℃ | ||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  CHESS CHESS  / Beamline: A1 / Wavelength: 0.908 / Beamline: A1 / Wavelength: 0.908 |
| Detector | Type: PRINCETON 1K / Detector: CCD / Date: Oct 15, 1996 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.908 Å / Relative weight: 1 |
| Reflection | Resolution: 2.35→50 Å / Num. all: 12843 / Num. obs: 12188 / % possible obs: 95 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 4.3 % / Biso Wilson estimate: 22.4 Å2 / Rmerge(I) obs: 0.088 / Net I/σ(I): 13 |
| Reflection shell | Resolution: 2.35→2.43 Å / Redundancy: 2.1 % / Rmerge(I) obs: 0.113 / % possible all: 86 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2.35→20 Å / Rfactor Rfree error: 0.009 / Data cutoff high absF: 100000 / Data cutoff low absF: 0.1 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 999.999 / Stereochemistry target values: ENGH AND HUBER
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 10.1 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.35→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.35→2.43 Å / Rfactor Rfree error: 0.033 / Total num. of bins used: 10
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: X-PLOR(ONLINE) / Version: 3.851 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS σ(F): 2 / % reflection Rfree: 7.2 % / Rfactor obs: 0.206 / Rfactor Rfree: 0.277 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 10.1 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor Rfree: 0.387 / % reflection Rfree: 12.6 % / Rfactor Rwork: 0.216 |
 Movie
Movie Controller
Controller












 PDBj
PDBj









