[English] 日本語
 Yorodumi
Yorodumi- PDB-1q11: Crystal structure of an active fragment of human tyrosyl-tRNA syn... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1q11 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of an active fragment of human tyrosyl-tRNA synthetase with tyrosinol | ||||||
 Components Components | Tyrosyl-tRNA synthetase | ||||||
 Keywords Keywords | LIGASE / Rossmann fold domain containing the active site / anticodon recognition domain / substrate analog tyrosinol co-crystalized | ||||||
| Function / homology |  Function and homology information Function and homology informationinterleukin-8 receptor binding / tyrosyl-tRNA aminoacylation / tyrosine-tRNA ligase / tyrosine-tRNA ligase activity / Cytosolic tRNA aminoacylation / response to starvation / small molecule binding / tRNA binding / nuclear body / apoptotic process ...interleukin-8 receptor binding / tyrosyl-tRNA aminoacylation / tyrosine-tRNA ligase / tyrosine-tRNA ligase activity / Cytosolic tRNA aminoacylation / response to starvation / small molecule binding / tRNA binding / nuclear body / apoptotic process / extracellular space / RNA binding / ATP binding / nucleus / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.6 Å MOLECULAR REPLACEMENT / Resolution: 1.6 Å | ||||||
 Authors Authors | Yang, X.-L. / Schimmel, P. / Ribas de Pouplana, L. | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2003 Journal: Proc.Natl.Acad.Sci.USA / Year: 2003Title: Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains Authors: Yang, X.-L. / Otero, F.J. / Skene, R.J. / McRee, D.E. / Schimmel, P. / Ribas De Pouplana, L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1q11.cif.gz 1q11.cif.gz | 88.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1q11.ent.gz pdb1q11.ent.gz | 65.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1q11.json.gz 1q11.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/q1/1q11 https://data.pdbj.org/pub/pdb/validation_reports/q1/1q11 ftp://data.pdbj.org/pub/pdb/validation_reports/q1/1q11 ftp://data.pdbj.org/pub/pdb/validation_reports/q1/1q11 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1r6tC  1n3lS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
| ||||||||
| Details | asymmetric unit contains a monomer of the biologically dimer |
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 42133.559 Da / Num. of mol.: 1 / Fragment: mini TyrRS Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Plasmid: PET20b / Production host: Homo sapiens (human) / Plasmid: PET20b / Production host:  |
|---|
-Non-polymers , 5 types, 353 molecules 

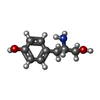






| #2: Chemical | ChemComp-PO4 / | ||
|---|---|---|---|
| #3: Chemical | ChemComp-K / | ||
| #4: Chemical | ChemComp-TYE / | ||
| #5: Chemical | | #6: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.58 Å3/Da / Density % sol: 52.41 % | |||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 279 K / Method: vapor diffusion, sitting drop / pH: 6.9 Details: (NH4)2SO4, NAH2PO4, K2HPO4, ACETONE, pH 6.9, VAPOR DIFFUSION, SITTING DROP, temperature 279K | |||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 4 ℃ / pH: 7.5 / Method: vapor diffusion, sitting drop | |||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X26C / Wavelength: 1.28 Å / Beamline: X26C / Wavelength: 1.28 Å |
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Nov 19, 2002 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.28 Å / Relative weight: 1 |
| Reflection | Resolution: 1.6→50 Å / Num. all: 58862 / Num. obs: 53993 / % possible obs: 91.7 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 6.3 % / Biso Wilson estimate: 16.2 Å2 / Rmerge(I) obs: 0.062 / Rsym value: 0.062 / Net I/σ(I): 16.2 |
| Reflection shell | Resolution: 1.6→1.66 Å / Rmerge(I) obs: 0.301 / Mean I/σ(I) obs: 3.8 / Rsym value: 0.301 / % possible all: 64.5 |
| Reflection | *PLUS Num. obs: 55381 / % possible obs: 93.8 % / Redundancy: 6.7 % |
| Reflection shell | *PLUS % possible obs: 64.5 % |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 1N3L, mini TyrRS alone Resolution: 1.6→50 Å / σ(F): 0 / Stereochemistry target values: Engh & Huber
| |||||||||||||||||||||||||
| Displacement parameters |
| |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.6→50 Å
| |||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||
| Refinement | *PLUS % reflection Rfree: 5 % / Rfactor Rfree: 0.217 | |||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller













 PDBj
PDBj




