[English] 日本語
 Yorodumi
Yorodumi- PDB-1nem: Saccharide-RNA recognition in the neomycin B / RNA aptamer complex -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1nem | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
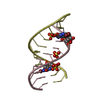
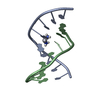
| Title | Saccharide-RNA recognition in the neomycin B / RNA aptamer complex | |||||||||||||||||||||
 Components Components | 5'-R(* Keywords KeywordsRNA / RNA APTAMER / AMINOGLYCOSIDE / ANTIBIOTIC | Function / homology | 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranose / 2-DEOXY-D-STREPTAMINE / RNA / RNA (> 10) |  Function and homology information Function and homology informationMethod | SOLUTION NMR / DISTANCE RESTRAINED MOLECULAR DYNAMICS |  Authors AuthorsJiang, L. / Majumdar, A. / Hu, W. / Jaishree, T.J. / Xu, W. / Patel, D.J. |  Citation Citation Journal: Structure Fold.Des. / Year: 1999 Journal: Structure Fold.Des. / Year: 1999Title: Saccharide-RNA recognition in a complex formed between neomycin B and an RNA aptamer Authors: Jiang, L. / Majumdar, A. / Hu, W. / Jaishree, T.J. / Xu, W. / Patel, D.J. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1nem.cif.gz 1nem.cif.gz | 165.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1nem.ent.gz pdb1nem.ent.gz | 134.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1nem.json.gz 1nem.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ne/1nem https://data.pdbj.org/pub/pdb/validation_reports/ne/1nem ftp://data.pdbj.org/pub/pdb/validation_reports/ne/1nem ftp://data.pdbj.org/pub/pdb/validation_reports/ne/1nem | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
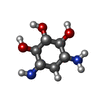
| #1: RNA chain | Mass: 7459.481 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: Complexed with neomycin B; hetero groups BDG (ring A), NEB (ring B), RIB (ring C), and NED (ring D) |
|---|---|
| #2: Polysaccharide | 2,6-diamino-2,6-dideoxy-beta-L-idopyranose-(1-3)-beta-D-ribofuranose Source method: isolated from a genetically manipulated source |
| #3: Sugar | ChemComp-BDG / |
| #4: Chemical | ChemComp-NEB / |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|---|
| NMR details | Text: EXPERIMENTS CONDUCTED IN D2O: 2D NOESY (50MS,120MS,200MS,300MS) PHASE-SENSITIVE COSY, TOCSY, (1H)- (13)C HSQC 3D (1H)-(13)C NOESY-HMQC (120MS, 300MS) 3D HCCH- COSY 3D HCCH-TOCSY 2D (1H)-(13)C ...Text: EXPERIMENTS CONDUCTED IN D2O: 2D NOESY (50MS,120MS,200MS,300MS) PHASE-SENSITIVE COSY, TOCSY, (1H)- (13)C HSQC 3D (1H)-(13)C NOESY-HMQC (120MS, 300MS) 3D HCCH- COSY 3D HCCH-TOCSY 2D (1H)-(13)C HCCH-TOCSY (65MS) 2D (1H)-(31)P CORRELATION 3D (1H)-(13)C-(31)P CORRELATION |
- Sample preparation
Sample preparation
| Sample conditions | Ionic strength: 10mM NA3PO4 / pH: 6.1 / Pressure: 1 atm / Temperature: 298 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
- Processing
Processing
| NMR software | Name:  X-PLOR / Version: 3.1 / Developer: BRUNGER / Classification: refinement X-PLOR / Version: 3.1 / Developer: BRUNGER / Classification: refinement |
|---|---|
| Refinement | Method: DISTANCE RESTRAINED MOLECULAR DYNAMICS / Software ordinal: 1 |
| NMR ensemble | Conformer selection criteria: LEAST RESTRAINT VIOLATION / Conformers calculated total number: 100 / Conformers submitted total number: 9 |
 Movie
Movie Controller
Controller






 PDBj
PDBj