+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1bvj | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
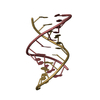
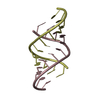
| Title | HIV-1 RNA A-RICH HAIRPIN LOOP | ||||||||||||||||||||
 Components Components | RNA (5'-R(P* Keywords KeywordsRNA / HIV-1 GENOMIC RNA / REVERSE TRANSCRIPTASE INITIATION / RNA HAIRPIN LOOP | Function / homology | : / RNA / RNA (> 10) |  Function and homology information Function and homology informationBiological species |   Human immunodeficiency virus type 1 Human immunodeficiency virus type 1Method | SOLUTION NMR / simulated annealing |  Authors AuthorsPuglisi, E.V. / Puglisi, J.D. |  Citation Citation Journal: Nat.Struct.Biol. / Year: 1998 Journal: Nat.Struct.Biol. / Year: 1998Title: HIV-1 A-rich RNA loop mimics the tRNA anticodon structure. Authors: Puglisi, E.V. / Puglisi, J.D. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1bvj.cif.gz 1bvj.cif.gz | 296.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1bvj.ent.gz pdb1bvj.ent.gz | 248.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1bvj.json.gz 1bvj.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bv/1bvj https://data.pdbj.org/pub/pdb/validation_reports/bv/1bvj ftp://data.pdbj.org/pub/pdb/validation_reports/bv/1bvj ftp://data.pdbj.org/pub/pdb/validation_reports/bv/1bvj | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 7402.472 Da / Num. of mol.: 1 / Fragment: A-RICH HAIRPIN LOOP / Source method: isolated from a natural source Source: (natural)  Human immunodeficiency virus type 1 (MAL ISOLATE) Human immunodeficiency virus type 1 (MAL ISOLATE)Genus: Lentivirus / Species: Human immunodeficiency virus 1 / Strain: MAL ISOLATE / References: GenBank: AF096656 |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||
| NMR details | Text: THE STRUCTURE WAS DETERMINED USING TRIPLE RESONANCE HCN AND HCP EXPERIMENTS ON 15N,13C-LABELED RNA. |
- Sample preparation
Sample preparation
| Sample conditions | Ionic strength: 10mM NA PHOSPHATE / pH: 6.4 / Temperature: 283 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Varian INOVA / Manufacturer: Varian / Model: INOVA / Field strength: 500 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 / Details: SEE REFERENCE ABOVE | ||||||||||||
| NMR ensemble | Conformer selection criteria: LOWEST TOTAL AND RESTRAINT VIOLATION ENERGIES Conformers calculated total number: 160 / Conformers submitted total number: 21 |
 Movie
Movie Controller
Controller







 PDBj
PDBj





























