+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1mag | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | GRAMICIDIN A IN HYDRATED DMPC BILAYERS, SOLID STATE NMR | |||||||||
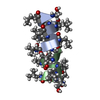
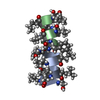
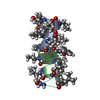
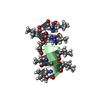
 Components Components | GRAMICIDIN A | |||||||||
 Keywords Keywords | ANTIBIOTIC / GRAMICIDIN / ANTIFUNGAL / ANTIBACTERIAL / MEMBRANE ION CHANNEL / LINEAR GRAMICIDIN / ORIENTED BILAYERS | |||||||||
| Function / homology | GRAMICIDIN A / :  Function and homology information Function and homology information | |||||||||
| Biological species |  BREVIBACILLUS BREVIS (bacteria) BREVIBACILLUS BREVIS (bacteria) | |||||||||
| Method | SOLID-STATE NMR / simulated annealing | |||||||||
 Authors Authors | Ketchem, R.R. / Roux, B. / Cross, T.A. | |||||||||
 Citation Citation |  Journal: J.Biomol.NMR / Year: 1996 Journal: J.Biomol.NMR / Year: 1996Title: Macromolecular Structural Elucidation with Solid-State NMR-Derived Orientational Constraints. Authors: Ketchem, R.R. / Lee, K.C. / Huo, S. / Cross, T.A. #1:  Journal: Biological Membranes: A Molecular Perspective from Computation and Experiment Journal: Biological Membranes: A Molecular Perspective from Computation and ExperimentYear: 1996 Title: Computational Refinement Through Solid State NMR and Energy Constraints of a Membrane Bound Polypeptide Authors: Ketchem, R.R. / Roux, B. / Cross, T.A. #2: Journal: Biochemistry / Year: 1995 Title: Lipid-Peptide Interface: Valine Conformation and Dynamics in the Gramicidin Channel Authors: Lee, K.C. / Huo, S. / Cross, T.A. #3: Journal: Biochemistry / Year: 1995 Title: Tryptophan Dynamics and Structural Refinement in a Lipid Bilayer Environment: Solid State NMR of the Gramicidin Channel Authors: Hu, W. / Lazo, N.D. / Cross, T.A. #4: Journal: Science / Year: 1993 Title: High-Resolution Conformation of Gramicidin a in a Lipid Bilayer by Solid-State NMR Authors: Ketchem, R.R. / Hu, W. / Cross, T.A. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1mag.cif.gz 1mag.cif.gz | 20 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1mag.ent.gz pdb1mag.ent.gz | 15.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1mag.json.gz 1mag.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1mag_validation.pdf.gz 1mag_validation.pdf.gz | 334.4 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1mag_full_validation.pdf.gz 1mag_full_validation.pdf.gz | 334.2 KB | Display | |
| Data in XML |  1mag_validation.xml.gz 1mag_validation.xml.gz | 2.1 KB | Display | |
| Data in CIF |  1mag_validation.cif.gz 1mag_validation.cif.gz | 2.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ma/1mag https://data.pdbj.org/pub/pdb/validation_reports/ma/1mag ftp://data.pdbj.org/pub/pdb/validation_reports/ma/1mag ftp://data.pdbj.org/pub/pdb/validation_reports/ma/1mag | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 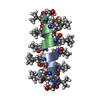
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
-Experimental details
-Experiment
| Experiment | Method: SOLID-STATE NMR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Sample conditions | pH: 7 / Temperature: 303 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: CHEMAGNETICS CMX / Manufacturer: CHEMAGNETICS / Model: CMX / Field strength: 400 MHz |
|---|
- Processing
Processing
| NMR software |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 Details: CHARMM23 STRUCTURE REFINED WITH TORC (TOTAL REFINEMENT OF CONSTRAINTS), DEVELOPED IN AUTHORS' LAB. TORC RUNS AS A MODULE WITHIN CHARMM AND UTILIZES A SIMULATED ANNEALING PROTOCOL TO REFINE ...Details: CHARMM23 STRUCTURE REFINED WITH TORC (TOTAL REFINEMENT OF CONSTRAINTS), DEVELOPED IN AUTHORS' LAB. TORC RUNS AS A MODULE WITHIN CHARMM AND UTILIZES A SIMULATED ANNEALING PROTOCOL TO REFINE THE STRUCTURE AGAINST BOTH THE SOLID STATE NMR DATA (INCLUDING SOLID STATE NMR DERIVED ORIENTATIONAL CONSTRAINTS) AND THE CHARMM ENERGY. | |||||||||
| NMR ensemble | Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller














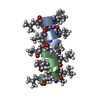
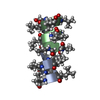
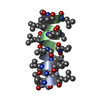

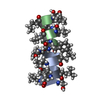
 PDBj
PDBj

