[English] 日本語
 Yorodumi
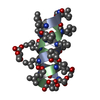
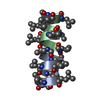
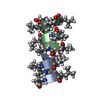
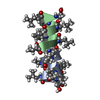
Yorodumi- PDB-1tkq: SOLUTION STRUCTURE OF A LINKED UNSYMMETRIC GRAMICIDIN IN A MEMBRA... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1tkq | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SOLUTION STRUCTURE OF A LINKED UNSYMMETRIC GRAMICIDIN IN A MEMBRANE-ISOELECTRICAL SOLVENTS MIXTURE IN THE PRESENCE OF CsCl | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | ANTIBIOTIC / GRAMICIDIN / ANTIFUNGAL / ANTIBACTERIAL / MEMBRANE ION CHANNEL / LINEAR GRAMICIDIN | |||||||||
| Function / homology | MINI-GRAMICIDIN A - GRAMICIDIN A DIMER / SUCCINIC ACID / :  Function and homology information Function and homology information | |||||||||
| Biological species |  BREVIBACILLUS BREVIS (bacteria) BREVIBACILLUS BREVIS (bacteria) | |||||||||
| Method | SOLUTION NMR / simulated annealing | |||||||||
| Model type details | MINIMIZED AVERAGE | |||||||||
 Authors Authors | Xie, X. / Al-Momani, L. / Bockelmann, D. / Griesinger, C. / Koert, U. | |||||||||
 Citation Citation |  Journal: FEBS J. / Year: 2005 Journal: FEBS J. / Year: 2005Title: An Asymmetric Ion Channel Derived from Gramicidin A. Synthesis, Function and NMR Structure. Authors: Xie, X. / Al-Momani, L. / Reiss, P. / Griesinger, C. / Koert, U. #1: Journal: Science / Year: 1993 Title: High-Resolution Conformation of Gramicidin a in a Lipid Bilayer by Solid-State NMR Authors: Ketchem, R.R. / Hu, W. / Cross, T.A. #2:  Journal: J.Mol.Biol. / Year: 1996 Journal: J.Mol.Biol. / Year: 1996Title: Solution Structure of a Parallel Left-Handed Double-Helical Gramicidin-A Determined by 2D 1H NMR Authors: Chen, Y. / Tucker, A. / Wallace, B.A. #3:  Journal: Angew.Chem.Int.Ed.Engl. / Year: 2002 Journal: Angew.Chem.Int.Ed.Engl. / Year: 2002Title: Cation Control in Functional Helical Programming: Structures of a D,L-Peptide Ion Channel Authors: Arndt, H.-D. / Bockelmann, D. / Knoll, A. / Lamberth, S. / Griesinger, C. / Koert, U. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1tkq.cif.gz 1tkq.cif.gz | 18.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1tkq.ent.gz pdb1tkq.ent.gz | 13.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1tkq.json.gz 1tkq.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/tk/1tkq https://data.pdbj.org/pub/pdb/validation_reports/tk/1tkq ftp://data.pdbj.org/pub/pdb/validation_reports/tk/1tkq ftp://data.pdbj.org/pub/pdb/validation_reports/tk/1tkq | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide |   Type: Polypeptide / Class: Antibiotic / Mass: 1470.798 Da / Num. of mol.: 1 / Source method: obtained synthetically Type: Polypeptide / Class: Antibiotic / Mass: 1470.798 Da / Num. of mol.: 1 / Source method: obtained syntheticallyDetails: THE N-TERMINI OF THE TWO PEPTIDES, EACH A TRUNCATED GRAMICIDIN A, WERE LINKED BY A SUCCINIC ACID IN A HEAD-TO-HEAD MANNER. Source: (synth.)  BREVIBACILLUS BREVIS (bacteria) BREVIBACILLUS BREVIS (bacteria)References: NOR: NOR00243, MINI-GRAMICIDIN A - GRAMICIDIN A DIMER |
|---|---|
| #2: Protein/peptide |   Type: Polypeptide / Class: Antibiotic / Mass: 1811.216 Da / Num. of mol.: 1 / Source method: obtained synthetically Type: Polypeptide / Class: Antibiotic / Mass: 1811.216 Da / Num. of mol.: 1 / Source method: obtained syntheticallyDetails: THE N-TERMINI OF THE TWO PEPTIDES, EACH A TRUNCATED GRAMICIDIN A, WERE LINKED BY A SUCCINIC ACID IN A HEAD-TO-HEAD MANNER. Source: (synth.)  BREVIBACILLUS BREVIS (bacteria) BREVIBACILLUS BREVIS (bacteria)References: NOR: NOR00243, MINI-GRAMICIDIN A - GRAMICIDIN A DIMER |
| #3: Chemical | ChemComp-SIN /   Type: Polypeptide / Class: Antibiotic / Mass: 118.088 Da / Num. of mol.: 1 / Source method: obtained synthetically / Formula: C4H6O4 Type: Polypeptide / Class: Antibiotic / Mass: 118.088 Da / Num. of mol.: 1 / Source method: obtained synthetically / Formula: C4H6O4Details: THE N-TERMINI OF THE TWO PEPTIDES, EACH A TRUNCATED GRAMICIDIN A, WERE LINKED BY A SUCCINIC ACID IN A HEAD-TO-HEAD MANNER. References: MINI-GRAMICIDIN A - GRAMICIDIN A DIMER |
| Compound details | GRAMICIDIN IS A HETEROGENEOUS MIXTURE OF SEVERAL COMPOUNDS INCLUDING GRAMICIDIN A, B AND C WHICH ...GRAMICIDIN |
| Has protein modification | N |
| Sequence details | BOTH OF THE C-TERMINI HAVE ETHANOLAMINE ATTACHED WITH THE CAPPING GROUP T-BUTYLDIPHENYLSILYL, WHICH ...BOTH OF THE C-TERMINI HAVE ETHANOLAMI |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||
| NMR details | Text: VARIABLE TEMPERATURE EXPERIMENTS WERE PERFORMED TO HAVE THE TEMPERATURE DEPENDENCE OF NH CHEMICAL SHIFTS, THEREFORE PROVIDE EVIDENCE FOR HYDROGEN BONDING. |
- Sample preparation
Sample preparation
| Details | Contents: 3MM SATURATED WITH CSCL |
|---|---|
| Sample conditions | Temperature: 293 K |
-NMR measurement
| NMR spectrometer | Type: Bruker AVANCE / Manufacturer: Bruker / Model: AVANCE / Field strength: 800 MHz |
|---|
- Processing
Processing
| NMR software |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 Details: THE STRUCTURES ARE BASED ON 160 NOE- DERIVED DISTANCE CONSTRAINTS, 39 DIHEDRAL ANGLE RESTRAINTS, AND 40 DISTANCE RESTRAINTS FROM HYDROGEN BONDS | |||||||||
| NMR ensemble | Conformer selection criteria: MINIMIZED AVERAGE STRUCTURE OF 11 STRUCTURES WITH THE LOWEST TARGET FUNCTION Conformers calculated total number: 50 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller


















 PDBj
PDBj