+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1gmk | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | GRAMICIDIN/KSCN COMPLEX | |||||||||
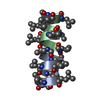
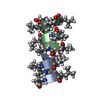
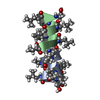
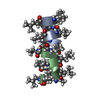
 Components Components | GRAMICIDIN A | |||||||||
 Keywords Keywords | ANTIBIOTIC / GRAMICIDIN / ANTIFUNGAL / ANTIBACTERIAL / MEMBRANE ION CHANNEL / LINEAR GRAMICIDIN / DOUBLE HELIX | |||||||||
| Function / homology | GRAMICIDIN A / : / METHANOL / THIOCYANATE ION / :  Function and homology information Function and homology information | |||||||||
| Biological species |  BREVIBACILLUS BREVIS (bacteria) BREVIBACILLUS BREVIS (bacteria) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.5 Å MOLECULAR REPLACEMENT / Resolution: 2.5 Å | |||||||||
 Authors Authors | Doyle, D.A. / Wallace, B.A. | |||||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1997 Journal: J.Mol.Biol. / Year: 1997Title: Crystal Structure of the Gramicidin/Potassium Thiocyanate Complex. Authors: Doyle, D.A. / Wallace, B.A. #1:  Journal: Biophys.J. / Year: 1994 Journal: Biophys.J. / Year: 1994Title: The Structure of the Gramicidin/Kscn Complex Authors: Doyle, D.A. / Wallace, B.A. #2:  Journal: Ann.N.Y.Acad.Sci. / Year: 1984 Journal: Ann.N.Y.Acad.Sci. / Year: 1984Title: Crystalline Ion Complexes of Gramicidin A Authors: Kimball, M.R. / Wallace, B.A. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1gmk.cif.gz 1gmk.cif.gz | 27.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1gmk.ent.gz pdb1gmk.ent.gz | 21 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1gmk.json.gz 1gmk.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gm/1gmk https://data.pdbj.org/pub/pdb/validation_reports/gm/1gmk ftp://data.pdbj.org/pub/pdb/validation_reports/gm/1gmk ftp://data.pdbj.org/pub/pdb/validation_reports/gm/1gmk | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 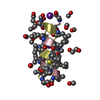
| ||||||||
| 2 | 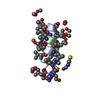
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein/peptide |   Type: Polypeptide / Class: Antibiotic / Mass: 1882.294 Da / Num. of mol.: 4 / Source method: isolated from a natural source Type: Polypeptide / Class: Antibiotic / Mass: 1882.294 Da / Num. of mol.: 4 / Source method: isolated from a natural sourceDetails: GRAMICIDIN A IS A HEXADECAMERIC HELICAL PEPTIDE WITH ALTERNATING D,L CHARACTERISTICS. THE N-TERM IS FORMYLATED (RESIDUE 0). THE C-TERM IS CAPPED WITH ETHANOLAMINE (RESIDUE 16). Source: (natural)  BREVIBACILLUS BREVIS (bacteria) / References: NOR: NOR00243, GRAMICIDIN A BREVIBACILLUS BREVIS (bacteria) / References: NOR: NOR00243, GRAMICIDIN A#2: Chemical | ChemComp-K / #3: Chemical | ChemComp-SCN / #4: Chemical | ChemComp-MOH / Compound details | GRAMICIDIN IS A HETEROGENEOUS MIXTURE OF SEVERAL COMPOUNDS INCLUDING GRAMICIDIN A, B AND C WHICH ...GRAMICIDIN | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Description: DATA COLLECTED AT ROOM TEMPERATURE WITH CRYSTAL IN METHANOL. | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Details: CRYSTALLIZED IN METHANOL | ||||||||||||||||||||
| Crystal grow | *PLUS Method: unknown | ||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 293 K |
|---|---|
| Diffraction source | Source: SEALED TUBE / Type: ENRAF-NONIUS / Wavelength: 1.5418 |
| Detector | Type: ENRAF-NONIUS FAST / Detector: DIFFRACTOMETER / Date: Oct 11, 1991 |
| Radiation | Monochromator: NI FILTER / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.5→10 Å / Num. obs: 5476 / % possible obs: 73.3 % / Observed criterion σ(I): 3 / Redundancy: 2 % / Rmerge(I) obs: 0.0325 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: GRAMICIDIN/CSCL COMPLEX, THEN GRAMICIDIN/CSSCN COMPLEX, BOTH WITHOUT IONS Resolution: 2.5→10 Å / σ(F): 3 Details: THE LOW TEMPERATURE FACTORS OF ATOMS N OF SCN 1, 2, 3, AND 4 ARE PROBABLY DUE TO POSITIONAL DISORDER OF THE THIOCYANATE MOLECULE WITH THE RESULT THAT OCCASIONALLY THE HEAVY SULFUR ATOM IS ...Details: THE LOW TEMPERATURE FACTORS OF ATOMS N OF SCN 1, 2, 3, AND 4 ARE PROBABLY DUE TO POSITIONAL DISORDER OF THE THIOCYANATE MOLECULE WITH THE RESULT THAT OCCASIONALLY THE HEAVY SULFUR ATOM IS SITUATED CLOSE TO THE PRESENT LOCATION OF THE NITROGEN. TWO METHANOL MOLECULES, MOH 5 AND MOH 19, WERE PLACED IN DENSITIES THAT ARE BELIEVED TO BELONG TO COMBINED DISORDERED CONFORMATIONS OF TRP A 15 AND TRP C 9 (MOH 19), AND TRP D 15 AND TRP A 9 (MOH 5).
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.5→10 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.1 / Classification: refinement X-PLOR / Version: 3.1 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller






















 PDBj
PDBj