[English] 日本語
 Yorodumi
Yorodumi- PDB-1kr3: Crystal Structure of the Metallo beta-Lactamase from Bacteroides ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1kr3 | ||||||
|---|---|---|---|---|---|---|---|
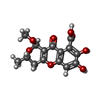
| Title | Crystal Structure of the Metallo beta-Lactamase from Bacteroides fragilis (CfiA) in Complex with the Tricyclic Inhibitor SB-236050. | ||||||
 Components Components | beta-Lactamase, type II | ||||||
 Keywords Keywords | HYDROLASE / beta sandwich / protein-inhibitor complex / metallo / beta-lactamase | ||||||
| Function / homology |  Function and homology information Function and homology informationantibiotic catabolic process / beta-lactamase activity / beta-lactamase / periplasmic space / response to antibiotic / zinc ion binding Similarity search - Function | ||||||
| Biological species |  Bacteroides fragilis (bacteria) Bacteroides fragilis (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.5 Å MOLECULAR REPLACEMENT / Resolution: 2.5 Å | ||||||
 Authors Authors | Payne, D.J. / Hueso-Rodrguez, J.A. / Boyd, H. / Concha, N.O. / Janson, C.A. / Gilpin, M. / Bateson, J.H. / Cheever, C. / Niconovich, N.L. / Pearson, S. ...Payne, D.J. / Hueso-Rodrguez, J.A. / Boyd, H. / Concha, N.O. / Janson, C.A. / Gilpin, M. / Bateson, J.H. / Cheever, C. / Niconovich, N.L. / Pearson, S. / Rittenhouse, S. / Tew, D. / Dez, E. / Prez, P. / de la Fuente, J. / Rees, M. / Rivera-Sagredo, A. | ||||||
 Citation Citation |  Journal: ANTIMICROB.AGENTS CHEMOTHER. / Year: 2002 Journal: ANTIMICROB.AGENTS CHEMOTHER. / Year: 2002Title: Identification of a series of tricyclic natural products as potent broad-spectrum inhibitors of metallo-beta-lactamases Authors: Payne, D.J. / Hueso-Rodrguez, J.A. / Boyd, H. / Concha, N.O. / Janson, C.A. / Gilpin, M. / Bateson, J.H. / Cheever, C. / Niconovich, N.L. / Pearson, S. / Rittenhouse, S. / Tew, D. / Dez, E. ...Authors: Payne, D.J. / Hueso-Rodrguez, J.A. / Boyd, H. / Concha, N.O. / Janson, C.A. / Gilpin, M. / Bateson, J.H. / Cheever, C. / Niconovich, N.L. / Pearson, S. / Rittenhouse, S. / Tew, D. / Dez, E. / Prez, P. / de la Fuente, J. / Rees, M. / Rivera-Sagredo, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1kr3.cif.gz 1kr3.cif.gz | 103.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1kr3.ent.gz pdb1kr3.ent.gz | 78.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1kr3.json.gz 1kr3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/kr/1kr3 https://data.pdbj.org/pub/pdb/validation_reports/kr/1kr3 ftp://data.pdbj.org/pub/pdb/validation_reports/kr/1kr3 ftp://data.pdbj.org/pub/pdb/validation_reports/kr/1kr3 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1hlkC  1znbS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 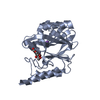
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | chain A and B do not form a functional dimer |
- Components
Components
| #1: Protein | Mass: 25358.627 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Bacteroides fragilis (bacteria) / Gene: CfiA / Plasmid: pET / Production host: Bacteroides fragilis (bacteria) / Gene: CfiA / Plasmid: pET / Production host:  #2: Chemical | ChemComp-ZN / #3: Chemical | #4: Chemical | #5: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.1 Å3/Da / Density % sol: 41.32 % | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop / pH: 6 Details: 32% PEG1000, 0.1M MES, 10microM ZnCl2, pH 6.0, VAPOR DIFFUSION, SITTING DROP, temperature 298K | ||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS pH: 7.5 | ||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 298 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: SIEMENS / Wavelength: 1.542 Å ROTATING ANODE / Type: SIEMENS / Wavelength: 1.542 Å |
| Detector | Type: SIEMENS / Detector: AREA DETECTOR / Date: Nov 1, 1997 / Details: Ni-filtered |
| Radiation | Monochromator: graphite / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.542 Å / Relative weight: 1 |
| Reflection | Resolution: 2.5→29 Å / Num. all: 15937 / Num. obs: 15937 / % possible obs: 75 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 2 / Redundancy: 1.9 % / Biso Wilson estimate: 20.6 Å2 / Rmerge(I) obs: 0.092 / Rsym value: 0.092 / Net I/σ(I): 9.3 |
| Reflection shell | Resolution: 2.5→2.66 Å / Redundancy: 1.9 % / Rmerge(I) obs: 0.23 / Mean I/σ(I) obs: 2.5 / Num. unique all: 1400 / Rsym value: 0.23 / % possible all: 56 |
| Reflection | *PLUS Lowest resolution: 50 Å / Num. obs: 10526 / % possible obs: 73.7 % |
| Reflection shell | *PLUS Highest resolution: 2.5 Å / Lowest resolution: 2.54 Å / % possible obs: 58 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: native CfiA (1ZNB) Resolution: 2.5→29.07 Å / Rfactor Rfree error: 0.007 / Data cutoff high absF: 813475.81 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 0 / Stereochemistry target values: Engh & Huber / Details: BULK SOLVENT MODEL USED
| ||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: MASK / Bsol: 60 Å2 / ksol: 0.33 e/Å3 | ||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 25.7 Å2
| ||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.5→29.07 Å
| ||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | NCS model details: CONSTRAINED MAIN CHAIN ATOMS | ||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.5→2.66 Å / Rfactor Rfree error: 0.026 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2.5 Å / Lowest resolution: 50 Å / % reflection Rfree: 10 % / Rfactor Rfree: 0.232 / Rfactor Rwork: 0.141 | ||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor Rwork: 0.22 |
 Movie
Movie Controller
Controller












 PDBj
PDBj