[English] 日本語
 Yorodumi
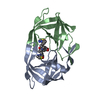
Yorodumi- PDB-1hih: COMPARATIVE ANALYSIS OF THE X-RAY STRUCTURES OF HIV-1 AND HIV-2 P... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1hih | ||||||
|---|---|---|---|---|---|---|---|
| Title | COMPARATIVE ANALYSIS OF THE X-RAY STRUCTURES OF HIV-1 AND HIV-2 PROTEASES IN COMPLEX WITH CGP 53820, A NOVEL PSEUDOSYMMETRIC INHIBITOR | ||||||
 Components Components | HIV-1 PROTEASE | ||||||
 Keywords Keywords | HYDROLASE (ASPARTIC PROTEINASE) / ASPARTATE PROTEASE / INHIBITED / HIV | ||||||
| Function / homology |  Function and homology information Function and homology informationHIV-1 retropepsin / symbiont-mediated activation of host apoptosis / retroviral ribonuclease H / exoribonuclease H / exoribonuclease H activity / DNA integration / viral genome integration into host DNA / RNA-directed DNA polymerase / establishment of integrated proviral latency / RNA stem-loop binding ...HIV-1 retropepsin / symbiont-mediated activation of host apoptosis / retroviral ribonuclease H / exoribonuclease H / exoribonuclease H activity / DNA integration / viral genome integration into host DNA / RNA-directed DNA polymerase / establishment of integrated proviral latency / RNA stem-loop binding / viral penetration into host nucleus / host multivesicular body / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / host cell / viral nucleocapsid / DNA recombination / DNA-directed DNA polymerase / aspartic-type endopeptidase activity / Hydrolases; Acting on ester bonds / DNA-directed DNA polymerase activity / symbiont-mediated suppression of host gene expression / viral translational frameshifting / symbiont entry into host cell / lipid binding / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / DNA binding / zinc ion binding / membrane Similarity search - Function | ||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 2.2 Å X-RAY DIFFRACTION / Resolution: 2.2 Å | ||||||
 Authors Authors | Priestle, J.P. / Gruetter, M.G. | ||||||
 Citation Citation |  Journal: Structure / Year: 1995 Journal: Structure / Year: 1995Title: Comparative analysis of the X-ray structures of HIV-1 and HIV-2 proteases in complex with CGP 53820, a novel pseudosymmetric inhibitor. Authors: Priestle, J.P. / Fassler, A. / Rosel, J. / Tintelnot-Blomley, M. / Strop, P. / Grutter, M.G. #1:  Journal: Bioorg.Med.Chem.Lett. / Year: 1993 Journal: Bioorg.Med.Chem.Lett. / Year: 1993Title: Novel Pseudosymmetric Inhibitors of HIV-1 Protease Authors: Fassler, A. / Rosel, J. / Tintelnot-Blomley, M. / Alteri, E. / Bold, G. / Lang, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1hih.cif.gz 1hih.cif.gz | 55.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1hih.ent.gz pdb1hih.ent.gz | 39.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1hih.json.gz 1hih.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hi/1hih https://data.pdbj.org/pub/pdb/validation_reports/hi/1hih ftp://data.pdbj.org/pub/pdb/validation_reports/hi/1hih ftp://data.pdbj.org/pub/pdb/validation_reports/hi/1hih | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Noncrystallographic symmetry (NCS) | NCS oper: (Code: given Matrix: (-0.9989, -0.04432, -0.015), Vector: Details | MTRIX THE TRANSFORMATIONS PRESENTED ON MTRIX RECORDS BELOW DESCRIBE NON-CRYSTALLOGRAPHIC RELATIONSHIPS AMONG THE VARIOUS DOMAINS IN THIS ENTRY. APPLYING THE APPROPRIATE MTRIX TRANSFORMATION TO THE RESIDUES LISTED FIRST WILL YIELD APPROXIMATE COORDINATES FOR THE RESIDUES LISTED SECOND. APPLIED TO TRANSFORMED TO MTRIX RESIDUES RESIDUES RMSD M1 A 1 .. A 99 B 1 .. B 99 0.677 | |
- Components
Components
| #1: Protein | Mass: 10803.756 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Human immunodeficiency virus 1 / Genus: Lentivirus / Cell line: S2 / Gene: POL / Plasmid: PT7Q10H / Gene (production host): POL / Production host: Human immunodeficiency virus 1 / Genus: Lentivirus / Cell line: S2 / Gene: POL / Plasmid: PT7Q10H / Gene (production host): POL / Production host:  References: UniProt: P03366, Hydrolases; Acting on peptide bonds (peptidases); Aspartic endopeptidases #2: Chemical | #3: Chemical | ChemComp-C20 / | #4: Water | ChemComp-HOH / | Compound details | COMPND MOLECULE: HIV-1 PROTEASE. BRU ISOLATE. | Has protein modification | N | Nonpolymer details | SOURCE 1 MOLECULE_NAME: CGP 53820. PSEUDOSYMMETRIC TRANSITION-STATE ANALOG. SOURCE 2 MOLECULE_NAME: ...SOURCE 1 MOLECULE_NAME: CGP 53820. PSEUDOSYMM | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.2 Å3/Da / Density % sol: 44.18 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal | *PLUS Density % sol: 45 % | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS pH: 5.4 / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction source | Wavelength: 1.5418 |
|---|---|
| Detector | Type: ENRAF-NONIUS FAST / Detector: DIFFRACTOMETER / Date: Aug 1, 1993 |
| Radiation | Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Num. obs: 10082 / % possible obs: 99.4 % / Observed criterion σ(I): 0 / Redundancy: 3.05 % / Rmerge(I) obs: 0.057 |
| Reflection | *PLUS Highest resolution: 2.2 Å / Num. measured all: 30789 / Rmerge(I) obs: 0.057 |
| Reflection shell | *PLUS Highest resolution: 2.2 Å / Lowest resolution: 2.26 Å / Rmerge(I) obs: 0.226 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2.2→6 Å / σ(F): 0 /
| ||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 23.8 Å2 | ||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.2→6 Å
| ||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||
| Software | *PLUS Name: TNT / Classification: refinement | ||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller





















 PDBj
PDBj