+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1gv4 | ||||||
|---|---|---|---|---|---|---|---|
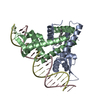
| Title | Murine apoptosis-inducing factor (AIF) | ||||||
 Components Components | PROGRAMED CELL DEATH PROTEIN 8 | ||||||
 Keywords Keywords | OXIDOREDUCTASE / FLAVOPROTEIN / FAD / NUCLEAR PROTEIN / APOPTOSI | ||||||
| Function / homology |  Function and homology information Function and homology informationelectron-transferring-flavoprotein dehydrogenase activity / regulation of apoptotic DNA fragmentation / Oxidoreductases; Acting on NADH or NADPH; With unknown physiological acceptors / mitochondrial respiratory chain complex assembly / poly-ADP-D-ribose binding / protein import into mitochondrial intermembrane space / NAD(P)H oxidase H2O2-forming activity / positive regulation of necroptotic process / NADH dehydrogenase activity / apoptotic mitochondrial changes ...electron-transferring-flavoprotein dehydrogenase activity / regulation of apoptotic DNA fragmentation / Oxidoreductases; Acting on NADH or NADPH; With unknown physiological acceptors / mitochondrial respiratory chain complex assembly / poly-ADP-D-ribose binding / protein import into mitochondrial intermembrane space / NAD(P)H oxidase H2O2-forming activity / positive regulation of necroptotic process / NADH dehydrogenase activity / apoptotic mitochondrial changes / intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress / FAD binding / mitochondrial intermembrane space / response to oxidative stress / neuron apoptotic process / mitochondrial outer membrane / protein dimerization activity / mitochondrial inner membrane / positive regulation of apoptotic process / perinuclear region of cytoplasm / mitochondrion / DNA binding / nucleus / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | ||||||
 Authors Authors | Mate, M.J. / Alzari, P.M. | ||||||
 Citation Citation |  Journal: Nat.Struct.Biol. / Year: 2002 Journal: Nat.Struct.Biol. / Year: 2002Title: The Crystal Structure of the Mouse Apoptosis-Inducing Factor Aif Authors: Mate, M.J. / Ortiz-Lombardia, M. / Alzari, P.M. / Boitel, B. / Haouz, A. / Tello, D. / Susin, S.A. / Penninger, J. / Kroemer, G. / Alzari, P.M. | ||||||
| History |
| ||||||
| Remark 700 | SHEET THE SHEET STRUCTURE OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN ... SHEET THE SHEET STRUCTURE OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN THE SHEET RECORDS BELOW, TWO SHEETS ARE DEFINED. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1gv4.cif.gz 1gv4.cif.gz | 209.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1gv4.ent.gz pdb1gv4.ent.gz | 164.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1gv4.json.gz 1gv4.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gv/1gv4 https://data.pdbj.org/pub/pdb/validation_reports/gv/1gv4 ftp://data.pdbj.org/pub/pdb/validation_reports/gv/1gv4 ftp://data.pdbj.org/pub/pdb/validation_reports/gv/1gv4 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1d7yS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Noncrystallographic symmetry (NCS) | NCS oper: (Code: given Matrix: (-0.99998, 0.00603, -0.00151), Vector: |
- Components
Components
| #1: Protein | Mass: 57571.426 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: Chemical | #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.3 Å3/Da / Density % sol: 43.5 % / Description: PHASE COMBINATION WITH MAD PHASES | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 7.75 Details: 18% PEG MONOMETHYL ETHER 5000, 100 MM MGCL2, 50 MM HEPES PH, pH 7.75 | ||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-4 / Wavelength: 0.93 / Beamline: ID14-4 / Wavelength: 0.93 |
| Detector | Detector: CCD / Date: Jun 15, 2000 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.93 Å / Relative weight: 1 |
| Reflection | Resolution: 2→20 Å / Num. obs: 62596 / % possible obs: 96.5 % / Redundancy: 2.9 % / Rmerge(I) obs: 0.11 / Net I/σ(I): 4.4 |
| Reflection shell | Resolution: 2→2.11 Å / Redundancy: 2.6 % / Rmerge(I) obs: 0.2 / Mean I/σ(I) obs: 2.5 / % possible all: 92.3 |
| Reflection | *PLUS Lowest resolution: 20 Å / % possible obs: 96.5 % / Rmerge(I) obs: 0.11 |
| Reflection shell | *PLUS % possible obs: 92.6 % / Rmerge(I) obs: 0.273 |
- Processing
Processing
| Software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1D7Y Resolution: 2→15 Å / SU B: 5.03 / SU ML: 0.14 / Cross valid method: THROUGHOUT / ESU R Free: 0.183 Details: DISORDERED RESIDUES WERE MODELED USING ROTAMERS DATA BASE AND HAVE OCC=0.00 IN THE PDB
| ||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→15 Å
| ||||||||||||||||||||
| Refinement | *PLUS Num. reflection obs: 71317 / % reflection Rfree: 5 % / Rfactor obs: 0.216 / Rfactor Rfree: 0.257 / Rfactor Rwork: 0.216 | ||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor Rfree: 0.339 / Rfactor Rwork: 0.24 / Rfactor obs: 0.24 |
 Movie
Movie Controller
Controller












 PDBj
PDBj





