[English] 日本語
 Yorodumi
Yorodumi- PDB-1c70: ALTERNATE BINDING SITE FOR THE P1-P3 GROUP OF A CLASS OF POTENT H... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1c70 | ||||||
|---|---|---|---|---|---|---|---|
| Title | ALTERNATE BINDING SITE FOR THE P1-P3 GROUP OF A CLASS OF POTENT HIV-1 PROTEASE INHIBITORS AS A RESULT OF CONCERTED STRUCTURAL CHANGE IN 80'S LOOP. | ||||||
 Components Components | PROTEIN (PROTEASE) | ||||||
 Keywords Keywords | HYDROLASE | ||||||
| Function / homology |  Function and homology information Function and homology informationhost multivesicular body / aspartic-type endopeptidase activity / virion membrane / proteolysis Similarity search - Function | ||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 2.5 Å X-RAY DIFFRACTION / Resolution: 2.5 Å | ||||||
 Authors Authors | Munshi, S. | ||||||
 Citation Citation |  Journal: Acta Crystallogr.,Sect.D / Year: 2000 Journal: Acta Crystallogr.,Sect.D / Year: 2000Title: An alternate binding site for the P1-P3 group of a class of potent HIV-1 protease inhibitors as a result of concerted structural change in the 80s loop of the protease. Authors: Munshi, S. / Chen, Z. / Yan, Y. / Li, Y. / Olsen, D.B. / Schock, H.B. / Galvin, B.B. / Dorsey, B. / Kuo, L.C. | ||||||
| History |
|
- Structure visualization
Structure visualization
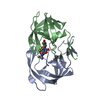
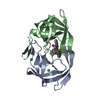
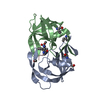
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1c70.cif.gz 1c70.cif.gz | 54.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1c70.ent.gz pdb1c70.ent.gz | 38.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1c70.json.gz 1c70.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/c7/1c70 https://data.pdbj.org/pub/pdb/validation_reports/c7/1c70 ftp://data.pdbj.org/pub/pdb/validation_reports/c7/1c70 ftp://data.pdbj.org/pub/pdb/validation_reports/c7/1c70 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | THERE IS ONE PROTEASE DIMER IN AN ASYMMETRICAL UNIT. THE TWO MOLECULES ARE LABELED AS CHAIN A AND CHAIN B. THERE IS ONE L-756,423 INHIBITOR MOLECULE LABELED AS L75. |
- Components
Components
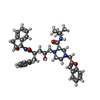
| #1: Protein | Mass: 10803.756 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: NY5 ISOLATE / Source: (gene. exp.)   Human immunodeficiency virus 1 / Genus: Lentivirus / Gene: NY5 ISOLATE / Production host: Human immunodeficiency virus 1 / Genus: Lentivirus / Gene: NY5 ISOLATE / Production host:  References: UniProt: O92103, Hydrolases; Acting on peptide bonds (peptidases); Aspartic endopeptidases #2: Chemical | ChemComp-L75 / | #3: Water | ChemComp-HOH / | Nonpolymer details | L-756,423 IS n-[2(r)-hydroxy-1(s)-indanyl]-2(r)-phenylmethyl -4(s)-hydroxy-5-[4-[2- ...L-756,423 IS n-[2(r)-hydroxy-1(s)-indanyl]-2(r)-phenylmethyl -4(s)-hydroxy-5-[4-[2-benzofuranylmethyl]-2(s)-[tert- butylaminocarbonyl]-piperazinyl]-pentaneamide | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.83 Å3/Da / Density % sol: 56.57 % Description: CRYSTALS OBTAINED BY CO-CRYSTALLIZATION AT PH 5.2, USING 0.6M NaCl AS PRECIPITATING AGENT IN 0.1M SODIUM ACETATE BUFFER. PROTEIN WAS AT 5.5 MG/ML CONCENTRATION. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS pH: 5 / Method: vapor diffusion, hanging dropDetails: inhibitor-protein mixture and the reservoir solution were mixed in a 1:1(v/v) ratio | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | Num. all: 33787 / Num. obs: 7292 / % possible obs: 86 % / Rmerge(I) obs: 0.074 |
| Reflection shell | Resolution: 2.5→2.59 Å / % possible all: 78 |
| Reflection | *PLUS Highest resolution: 2.5 Å / Lowest resolution: 20 Å / Num. measured all: 33787 |
| Reflection shell | *PLUS % possible obs: 78 % / Num. unique obs: 648 / Num. measured obs: 1291 / Rmerge(I) obs: 0.28 / Mean I/σ(I) obs: 1.8 |
- Processing
Processing
| Software | Name:  X-PLOR / Classification: refinement X-PLOR / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2.5→8 Å / Rfactor Rfree: 0.27 / Rfactor Rwork: 0.16 / Rfactor obs: 0.16 / σ(F): 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.5→8 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller






















 PDBj
PDBj