[English] 日本語
 Yorodumi
Yorodumi- PDB-1arz: ESCHERICHIA COLI DIHYDRODIPICOLINATE REDUCTASE IN COMPLEX WITH NA... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1arz | ||||||
|---|---|---|---|---|---|---|---|
| Title | ESCHERICHIA COLI DIHYDRODIPICOLINATE REDUCTASE IN COMPLEX WITH NADH AND 2,6 PYRIDINE DICARBOXYLATE | ||||||
 Components Components | DIHYDRODIPICOLINATE REDUCTASE | ||||||
 Keywords Keywords | OXIDOREDUCTASE / REDUCTASE / LYSINE BIOSYNTHESIS / NADH BINDING | ||||||
| Function / homology |  Function and homology information Function and homology information4-hydroxy-tetrahydrodipicolinate reductase / oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor / 4-hydroxy-tetrahydrodipicolinate reductase activity / diaminopimelate biosynthetic process / L-lysine biosynthetic process via diaminopimelate / amino acid biosynthetic process / NAD binding / NADP binding / identical protein binding / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.6 Å MOLECULAR REPLACEMENT / Resolution: 2.6 Å | ||||||
 Authors Authors | Scapin, G. / Reddy, S.G. / Zheng, R. / Blanchard, J.S. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1997 Journal: Biochemistry / Year: 1997Title: Three-dimensional structure of Escherichia coli dihydrodipicolinate reductase in complex with NADH and the inhibitor 2,6-pyridinedicarboxylate. Authors: Scapin, G. / Reddy, S.G. / Zheng, R. / Blanchard, J.S. #1:  Journal: Biochemistry / Year: 1995 Journal: Biochemistry / Year: 1995Title: Three-Dimensional Structure of Escherichia Coli Dihydrodipicolinate Reductase Authors: Scapin, G. / Blanchard, J.S. / Sacchettini, J.C. #2:  Journal: Biochemistry / Year: 1995 Journal: Biochemistry / Year: 1995Title: Expression, Purification, and Characterization of Escherichia Coli Dihydrodipicolinate Reductase Authors: Reddy, S.G. / Sacchettini, J.C. / Blanchard, J.S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1arz.cif.gz 1arz.cif.gz | 212.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1arz.ent.gz pdb1arz.ent.gz | 171.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1arz.json.gz 1arz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ar/1arz https://data.pdbj.org/pub/pdb/validation_reports/ar/1arz ftp://data.pdbj.org/pub/pdb/validation_reports/ar/1arz ftp://data.pdbj.org/pub/pdb/validation_reports/ar/1arz | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1dihS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
-Protein , 1 types, 4 molecules ABCD
| #1: Protein | Mass: 28793.631 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|
-Non-polymers , 5 types, 188 molecules 


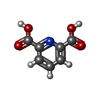





| #2: Chemical | ChemComp-PO4 / | ||||||
|---|---|---|---|---|---|---|---|
| #3: Chemical | | #4: Chemical | #5: Chemical | #6: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.42 Å3/Da / Density % sol: 50 % | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 7.5 Details: PROTEIN WAS CRYSTALLIZED FROM 23-26% PEG 8000, IN 160-180 MM POTASSIUM PHOSPHATE, 100 MM SODIUM CACODYLATE PH 7.5 | ||||||||||||||||||||||||||||||||||||||||||
| Crystal | *PLUS | ||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop / pH: 6.5 | ||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 294 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 |
| Detector | Type: SIEMENS / Detector: AREA DETECTOR / Date: Aug 1, 1996 |
| Radiation | Monochromator: NI FILTER / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.6→25 Å / Num. obs: 32245 / % possible obs: 90 % / Observed criterion σ(I): 0.5 / Redundancy: 3.7 % / Rmerge(I) obs: 0.04 / Rsym value: 0.081 / Net I/σ(I): 15.9 |
| Reflection shell | Resolution: 2.6→2.76 Å / Redundancy: 1.8 % / Rmerge(I) obs: 0.126 / Mean I/σ(I) obs: 2 / Rsym value: 0.269 / % possible all: 74 |
| Reflection | *PLUS Num. measured all: 119624 / Rmerge(I) obs: 0.081 |
| Reflection shell | *PLUS % possible obs: 74 % / Num. unique obs: 4365 / Num. measured obs: 8022 / Rmerge(I) obs: 0.269 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1DIH Resolution: 2.6→20 Å / Data cutoff high absF: 1000000 / Data cutoff low absF: 1.0E-5 / Cross valid method: THROUGHOUT / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 33.7 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati d res low obs: 20 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.6→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | NCS model details: RESTRAINTS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.6→2.72 Å / Total num. of bins used: 8
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.1 / Classification: refinement X-PLOR / Version: 3.1 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor Rwork: 0.32 |
 Movie
Movie Controller
Controller










 PDBj
PDBj





