+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9868 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rabbit Cav1.1-Verapamil Complex | |||||||||||||||||||||
 Map data Map data | EM map | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | Membrane protein complex modulated by FDA approved drug. / MEMBRANE PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationhigh voltage-gated calcium channel activity / L-type voltage-gated calcium channel complex / positive regulation of muscle contraction / regulation of release of sequestered calcium ion into cytosol / regulation of calcium ion transmembrane transport via high voltage-gated calcium channel / cellular response to caffeine / calcium ion import across plasma membrane / voltage-gated calcium channel activity / release of sequestered calcium ion into cytosol / T-tubule ...high voltage-gated calcium channel activity / L-type voltage-gated calcium channel complex / positive regulation of muscle contraction / regulation of release of sequestered calcium ion into cytosol / regulation of calcium ion transmembrane transport via high voltage-gated calcium channel / cellular response to caffeine / calcium ion import across plasma membrane / voltage-gated calcium channel activity / release of sequestered calcium ion into cytosol / T-tubule / muscle contraction / calcium channel regulator activity / sarcolemma / calcium ion transmembrane transport / transmembrane transporter binding / calmodulin binding / metal ion binding / plasma membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||||||||||||||
 Authors Authors | Zhao Y / Huang G | |||||||||||||||||||||
| Funding support |  China, 6 items China, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2019 Journal: Cell / Year: 2019Title: Molecular Basis for Ligand Modulation of a Mammalian Voltage-Gated Ca Channel. Authors: Yanyu Zhao / Gaoxingyu Huang / Jianping Wu / Qiurong Wu / Shuai Gao / Zhen Yan / Jianlin Lei / Nieng Yan /   Abstract: The L-type voltage-gated Ca (Ca) channels are modulated by various compounds exemplified by 1,4-dihydropyridines (DHP), benzothiazepines (BTZ), and phenylalkylamines (PAA), many of which have been ...The L-type voltage-gated Ca (Ca) channels are modulated by various compounds exemplified by 1,4-dihydropyridines (DHP), benzothiazepines (BTZ), and phenylalkylamines (PAA), many of which have been used for characterizing channel properties and for treatment of hypertension and other disorders. Here, we report the cryoelectron microscopy (cryo-EM) structures of Ca1.1 in complex with archetypal antagonistic drugs, nifedipine, diltiazem, and verapamil, at resolutions of 2.9 Å, 3.0 Å, and 2.7 Å, respectively, and with a DHP agonist Bay K 8644 at 2.8 Å. Diltiazem and verapamil traverse the central cavity of the pore domain, directly blocking ion permeation. Although nifedipine and Bay K 8644 occupy the same fenestration site at the interface of repeats III and IV, the coordination details support previous functional observations that Bay K 8644 is less favored in the inactivated state. These structures elucidate the modes of action of different Ca ligands and establish a framework for structure-guided drug discovery. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9868.map.gz emd_9868.map.gz | 116.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9868-v30.xml emd-9868-v30.xml emd-9868.xml emd-9868.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9868.png emd_9868.png | 13.6 KB | ||
| Filedesc metadata |  emd-9868.cif.gz emd-9868.cif.gz | 8.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9868 http://ftp.pdbj.org/pub/emdb/structures/EMD-9868 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9868 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9868 | HTTPS FTP |
-Related structure data
| Related structure data |  6jpaMC  9866C  9867C  9869C  6jp5C  6jp8C  6jpbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9868.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9868.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.091 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
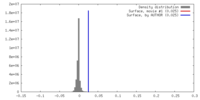
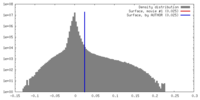
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Cav1.1-drug complex
+Supramolecule #1: Cav1.1-drug complex
+Supramolecule #2: Cav1.1-beta1a-alpha2-delta1-gamma complex
+Macromolecule #1: Voltage-dependent L-type calcium channel subunit alpha-1S
+Macromolecule #2: Voltage-dependent calcium channel gamma-1 subunit
+Macromolecule #3: Voltage-dependent L-type calcium channel subunit beta-1
+Macromolecule #4: Voltage-dependent L-type calcium channel subunit beta-1
+Macromolecule #5: Voltage-dependent calcium channel subunit alpha-2/delta-1
+Macromolecule #9: 2-acetamido-2-deoxy-beta-D-glucopyranose
+Macromolecule #10: CALCIUM ION
+Macromolecule #11: 1,2-Distearoyl-sn-glycerophosphoethanolamine
+Macromolecule #12: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
+Macromolecule #13: (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]sp...
+Macromolecule #14: (2S)-2-(3,4-dimethoxyphenyl)-5-{[2-(3,4-dimethoxyphenyl)ethyl](me...
+Macromolecule #15: ETHANOLAMINE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 433477 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)