[English] 日本語
 Yorodumi
Yorodumi- EMDB-9366: Atomic structures and deletion mutant reveal different capsid-bin... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9366 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Atomic structures and deletion mutant reveal different capsid-binding patterns and functional significance of tegument protein pp150 in murine and human cytomegaloviruses with implications for therapeutic development | ||||||||||||||||||
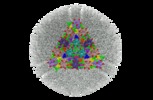
 Map data Map data | 3D reconstruction for MCMV capsid associated with pM32 tegument protein. Contour level=4.0 was estimated while showing the map using step=2 in Chimera. | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | cryoEM / beta-herpesvirus / murine cytomegalovirus / human cytomegalovirus / pp150 / pUL32 / pM32 / bacterial artificial chromosome-based mutagenesis / VIRUS | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationT=16 icosahedral viral capsid / viral tegument / viral capsid assembly / viral process / viral capsid / host cell nucleus / structural molecule activity / DNA binding Similarity search - Function | ||||||||||||||||||
| Biological species |  Murine cytomegalovirus (strain Smith) / Murine cytomegalovirus (strain Smith) /  Murid herpesvirus 1 (strain Smith) Murid herpesvirus 1 (strain Smith) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.0 Å | ||||||||||||||||||
 Authors Authors | Liu W / Dai XH | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2019 Journal: PLoS Pathog / Year: 2019Title: Atomic structures and deletion mutant reveal different capsid-binding patterns and functional significance of tegument protein pp150 in murine and human cytomegaloviruses with implications for ...Title: Atomic structures and deletion mutant reveal different capsid-binding patterns and functional significance of tegument protein pp150 in murine and human cytomegaloviruses with implications for therapeutic development. Authors: Wei Liu / Xinghong Dai / Jonathan Jih / Karen Chan / Phong Trang / Xuekui Yu / Rilwan Balogun / Ye Mei / Fenyong Liu / Z Hong Zhou /   Abstract: Cytomegalovirus (CMV) infection causes birth defects and life-threatening complications in immunosuppressed patients. Lack of vaccine and need for more effective drugs have driven widespread ongoing ...Cytomegalovirus (CMV) infection causes birth defects and life-threatening complications in immunosuppressed patients. Lack of vaccine and need for more effective drugs have driven widespread ongoing therapeutic development efforts against human CMV (HCMV), mostly using murine CMV (MCMV) as the model system for preclinical animal tests. The recent publication (Yu et al., 2017, DOI: 10.1126/science.aam6892) of an atomic model for HCMV capsid with associated tegument protein pp150 has infused impetus for rational design of novel vaccines and drugs, but the absence of high-resolution structural data on MCMV remains a significant knowledge gap in such development efforts. Here, by cryoEM with sub-particle reconstruction method, we have obtained the first atomic structure of MCMV capsid with associated pp150. Surprisingly, the capsid-binding patterns of pp150 differ between HCMV and MCMV despite their highly similar capsid structures. In MCMV, pp150 is absent on triplex Tc and exists as a "Λ"-shaped dimer on other triplexes, leading to only 260 groups of two pp150 subunits per capsid in contrast to 320 groups of three pp150 subunits each in a "Δ"-shaped fortifying configuration. Many more amino acids contribute to pp150-pp150 interactions in MCMV than in HCMV, making MCMV pp150 dimer inflexible thus incompatible to instigate triplex Tc-binding as observed in HCMV. While pp150 is essential in HCMV, our pp150-deletion mutant of MCMV remained viable though with attenuated infectivity and exhibiting defects in retaining viral genome. These results thus invalidate targeting pp150, but lend support to targeting capsid proteins, when using MCMV as a model for HCMV pathogenesis and therapeutic studies. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9366.map.gz emd_9366.map.gz | 1.4 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9366-v30.xml emd-9366-v30.xml emd-9366.xml emd-9366.xml | 26.7 KB 26.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9366.png emd_9366.png | 309.2 KB | ||
| Filedesc metadata |  emd-9366.cif.gz emd-9366.cif.gz | 8.1 KB | ||
| Others |  emd_9366_additional_1.map.gz emd_9366_additional_1.map.gz emd_9366_additional_2.map.gz emd_9366_additional_2.map.gz emd_9366_additional_3.map.gz emd_9366_additional_3.map.gz | 220.8 MB 219.3 MB 219.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9366 http://ftp.pdbj.org/pub/emdb/structures/EMD-9366 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9366 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9366 | HTTPS FTP |
-Related structure data
| Related structure data |  6nhjMC  9367C  9368C  9369C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9366.map.gz / Format: CCP4 / Size: 7.8 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9366.map.gz / Format: CCP4 / Size: 7.8 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D reconstruction for MCMV capsid associated with pM32 tegument protein. Contour level=4.0 was estimated while showing the map using step=2 in Chimera. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
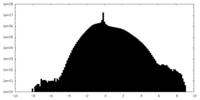
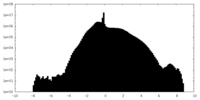
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.351 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
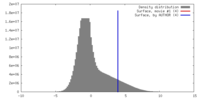
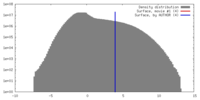
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Sub-particle reconstruction for MCMV 2-fold region
| File | emd_9366_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sub-particle reconstruction for MCMV 2-fold region | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sub-particle reconstruction for MCMV 3-fold region
| File | emd_9366_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sub-particle reconstruction for MCMV 3-fold region | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sub-particle reconstruction for MCMV 5-fold region
| File | emd_9366_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sub-particle reconstruction for MCMV 5-fold region | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Murid herpesvirus 1 (strain Smith)
| Entire | Name:  Murid herpesvirus 1 (strain Smith) Murid herpesvirus 1 (strain Smith) |
|---|---|
| Components |
|
-Supramolecule #1: Murid herpesvirus 1 (strain Smith)
| Supramolecule | Name: Murid herpesvirus 1 (strain Smith) / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10367 / Sci species name: Murid herpesvirus 1 (strain Smith) / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 16 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Murine cytomegalovirus (strain Smith) / Strain: Smith Murine cytomegalovirus (strain Smith) / Strain: Smith |
| Molecular weight | Theoretical: 151.673031 KDa |
| Sequence | String: MGENWTATEL LPKLDVPIDL LTHIKLSVGE EMFNNFRLYY GDDPERYNLS FEAIFGTYCN KIEWVTFLGT ALATAAHAIM FHDLNKMTT GKMLFYIQVP RVATGAGIPT SRQTTVMVSK YSEKSPITIP FEISAACLTH LKETFEDTLL DKLLNADAVN T VLRAVKNT ...String: MGENWTATEL LPKLDVPIDL LTHIKLSVGE EMFNNFRLYY GDDPERYNLS FEAIFGTYCN KIEWVTFLGT ALATAAHAIM FHDLNKMTT GKMLFYIQVP RVATGAGIPT SRQTTVMVSK YSEKSPITIP FEISAACLTH LKETFEDTLL DKLLNADAVN T VLRAVKNT ADAMERGLID TFLRVLLRHA PPCFVLRTLM EHGTIARRMA TRVQRANIAQ GFKSKMLATI FLLDRSRDRG QL TRYLDML TDCVTESILD NPETYTVGGG ERLAGVIVST HTVVQALLNA LGGSIRRTGV KTPASYGKFV LSKENAVTAI AHH AIMADF SQHADRIQQS SQKDLPESQF LDQRLTFTET QMDVLKVGER LVALEHLRKV YKNTDVQDPL ERDVELTFYF PVGL HVPSG RAYSTAENKI KLVDTAENQL PTTVYFYNKD RIPQRISHAE ALKTLCHPAI HDAGPCLEAF AQAGPPQGDD RVRAL CRRE FVREHMAHAT RRLVHFYQAR IDPPRTANEA KHDFSTKEFA KVDNYLLFTE LHPFFDFCFH TENGQVRPLC TPRIMV GNL PEALAPADFH DLRAKQALEL TKVRAPEGHE ATLQVLRASL TDHQYPELFY LIESLIHGDP AAFETGIELV TRCVNNY WR QRGLLAFANS YDMVRLIATR LGDGAVVPAA YTHYRNLLSI TRFVARTCEL TGLNGRLCDE PLLAYVSALH DPRLWPPF V QALPRNANLV RVVADDVPLD AAHIEERNPG TSDVARMIAM DQAEPLFVDA RRTSDEEMVA QKVYYLCLVP AVLNNHACG AGLNLKHLLV KLFYTKFFLT ADPDSLTAGE EALTNNPLLA ALVRDVATDE NVTANQAAEE LFHLVAHVPE NAQMLEIRAA LDPAQRHGA PSAGFESLQH VLYNGFCMTT VPKLLQEYLT VIPFHRFYSD PGLAATANHD IRVFLNDFPQ YQRCDGGFPL S PIFAHEYH HWHRTPFSCY SAACAHTLES VLTLAIMHHK MSPVSIAALS RMGLHPGFAL TVVRTDTFET DTLLYSTKAS TA VIINTPI VTKEDRDINT VFHVSQNINT VEMGLGYGAT TCTAHLRRVR SDMGSRMQDL FQVFPMHVYR NEDVDAWVRQ ATG ARRTEV LDSEAISILT FGRKTDKGGP ALLHGQRATC EVILTPVSAD LEFFRYPNNP RGRSSSMLGV DPYDEDAALA TLYD HTSPD PQTFVSTNNP WGSQRGSLGD VIYNTRNREK LGHNPSFYSP CAQFFTTDDI INANKTLFKT VEEYLNRSQD CIHGE TDLQ YICVEGTNSI VEKPCRFLQE ALTQHTGTTQ ALMESQLKGT SKLGLDETHY GNYSIGETIP LQQSILFNS UniProtKB: Major capsid protein |
-Macromolecule #2: Tegument protein
| Macromolecule | Name: Tegument protein / type: protein_or_peptide / ID: 2 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Murine cytomegalovirus (strain Smith) / Strain: Smith Murine cytomegalovirus (strain Smith) / Strain: Smith |
| Molecular weight | Theoretical: 78.749961 KDa |
| Sequence | String: MSARGRAAGD DGRQAELMAT LGFVRLSKSS VGKVKKFLNN LYDLKSINLC RHPRVIAECR GTDLSRETQL YNEMVLWLRY HEKLTARRP GHLPLLTRIR QDYDKLFGFV AARPELCGFD GLTEVNVFDD AVYGDDYVPR VDVFLRGLED LARCLCAQGP D KPARAVIM ...String: MSARGRAAGD DGRQAELMAT LGFVRLSKSS VGKVKKFLNN LYDLKSINLC RHPRVIAECR GTDLSRETQL YNEMVLWLRY HEKLTARRP GHLPLLTRIR QDYDKLFGFV AARPELCGFD GLTEVNVFDD AVYGDDYVPR VDVFLRGLED LARCLCAQGP D KPARAVIM GFINMRAEEV NRLMDNVRDA AERVLVYEVL DVRDPLNEDP SVLVHNRLVY LCRLAYAISK SWQTLSHMCL DR INSLRRR LILAFHDRPA FARVYARNAL ERPVDGTTAY NLLRRLEEDF LLFRNALRWG DPDWGLESEL ESEGDNSDAG SDL DLEDEE DDDDGGGPGG HDDESGGNRT PDPGMSLHDD TGIANTCLIG GDDEDCGGGS GRCLEFDPDS ERCLGVKMVN GRAL RWWNP TGIMVDNEAA VWIDEHGRVM DKPPPKELRK ASSDDGGNKK NPPPKKNVTP PVSGSNSVGG GVQTPSTASG KRTGK KKEG GGGYLLRSRS TDDDEVRKMK KDGTIDDRAD RELKMALQKA RESTADSDLS TILPRTEPLR KVAFVGDPVA AFGDTV RTT SSSKGFDDGP FTTQGASVLL PPPLGGPGST LTLPPDLPDL SGVLADEDVQ DSRYGKIPKS RTKKHPTFPE NYKQRPP PH NKDDEYYWDE TGDNMPVGED GGGVLEDLRK GLEGIDLKTG GGGSLQPPLS QQFAGSPFAG SDGDGGGLVK KSSSSH UniProtKB: Tegument protein |
-Macromolecule #3: Small capsomere-interacting protein
| Macromolecule | Name: Small capsomere-interacting protein / type: protein_or_peptide / ID: 3 / Number of copies: 16 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Murine cytomegalovirus (strain Smith) / Strain: Smith Murine cytomegalovirus (strain Smith) / Strain: Smith |
| Molecular weight | Theoretical: 9.845198 KDa |
| Sequence | String: MSTNVSSAAS GGGSSGGSSG ASSGGGGGGS GGSSKKEEER RKQFGANVLN LAPAMVAQPV ISTMIPKYMK MGGHEDKLAY QLDLLRMLS IAKKATVIQ UniProtKB: Small capsomere-interacting protein |
-Macromolecule #4: Minor capsid protein
| Macromolecule | Name: Minor capsid protein / type: protein_or_peptide / ID: 4 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Murine cytomegalovirus (strain Smith) / Strain: Smith Murine cytomegalovirus (strain Smith) / Strain: Smith |
| Molecular weight | Theoretical: 33.281289 KDa |
| Sequence | String: MANVSSFGPM KREVMEFDPE DPYKVSKMRK LERSIAKGYV YGADHQAITA RFFVRESLGE VEQKNLGVLM FRLDTGIEMP STVLVSLFF LSMVAENVSA ATKNTLAAIY GREGEAIRTW LRDGAWRLHR VVHPLGCTNS ITPGATCLIT CSMRGHSYNM L KTEIYPLL ...String: MANVSSFGPM KREVMEFDPE DPYKVSKMRK LERSIAKGYV YGADHQAITA RFFVRESLGE VEQKNLGVLM FRLDTGIEMP STVLVSLFF LSMVAENVSA ATKNTLAAIY GREGEAIRTW LRDGAWRLHR VVHPLGCTNS ITPGATCLIT CSMRGHSYNM L KTEIYPLL VPKEIYLDLD GESTDEIRFV YFVITYDYNS DRQGRPSAFV VVSRITHRHT LINVLRYRFR VSRFHFLNNS IS GYGPSTG CLGTLQRLGW FCSRDSRSGI VASRAGQLSV VKLEKFYVDV GPLVEFA UniProtKB: Minor capsid protein |
-Macromolecule #5: Triplex capsid protein 2
| Macromolecule | Name: Triplex capsid protein 2 / type: protein_or_peptide / ID: 5 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Murine cytomegalovirus (strain Smith) / Strain: Smith Murine cytomegalovirus (strain Smith) / Strain: Smith |
| Molecular weight | Theoretical: 34.660262 KDa |
| Sequence | String: METTVLVTFE QRLTTGDVGK LSRLIGAVIP IPYRHHLLGS SQVGLDAVVK DKTRDYSRMR ARMREMTLTI MRRVEGNQMI LGVPTHGQC YTIRNTGPVS WEKGDVLTTL PPVFSGEVTG LVSVSDWDLV LPWIVPMALA TEINQRMMML ALLSLDRSHE E VRAATAQL ...String: METTVLVTFE QRLTTGDVGK LSRLIGAVIP IPYRHHLLGS SQVGLDAVVK DKTRDYSRMR ARMREMTLTI MRRVEGNQMI LGVPTHGQC YTIRNTGPVS WEKGDVLTTL PPVFSGEVTG LVSVSDWDLV LPWIVPMALA TEINQRMMML ALLSLDRSHE E VRAATAQL RVVRYRDATL TLPEITIDDT VLIDMRNVCI SLSMIANLSS EVTLAYVRKL ALEDSNMLLM KCQEILGRRM PQ VGVGAGS SGDRNDPPAR SRTNYNITPT EELNKLTALF VMIRQITDVI SEQPAFLVCD VSPDDKSALC IYKG UniProtKB: Uncharacterized protein M85 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: PBS buffer, PH=7.4 |
|---|---|
| Grid | Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER / Details: The grids were manually plunged into the Ethane.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: AGFA SCIENTA FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Number real images: 2200 / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.0 µm / Calibrated defocus min: 1.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 47000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 200 |
|---|---|
| Output model |  PDB-6nhj: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)