[English] 日本語
 Yorodumi
Yorodumi- EMDB-6213: CryoEM reconstruction of Kaposi's sarcoma-associated herpesvirus ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6213 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM reconstruction of Kaposi's sarcoma-associated herpesvirus mutant with C-terminal half of SCP truncated (KSHV-SCPN86) | |||||||||
 Map data Map data | CryoEM reconstruction of KSHV-SCPN86 mutant. The capsid structure is essentially identical to that of wild-type KSHV. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | herpesvirus / gammaherpesvirus / the smallest capsid protein / cementing protein | |||||||||
| Biological species |   Human herpesvirus 8 Human herpesvirus 8 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.4 Å | |||||||||
 Authors Authors | Dai XH / Gong DY / Xiao YC / Wu TT / Sun R / Zhou ZH | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2015 Journal: Proc Natl Acad Sci U S A / Year: 2015Title: CryoEM and mutagenesis reveal that the smallest capsid protein cements and stabilizes Kaposi's sarcoma-associated herpesvirus capsid. Authors: Xinghong Dai / Danyang Gong / Yuchen Xiao / Ting-Ting Wu / Ren Sun / Z Hong Zhou /  Abstract: With just one eighth the size of the major capsid protein (MCP), the smallest capsid protein (SCP) of human tumor herpesviruses--Kaposi's sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus ...With just one eighth the size of the major capsid protein (MCP), the smallest capsid protein (SCP) of human tumor herpesviruses--Kaposi's sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus (EBV)--is vital to capsid assembly, yet its mechanism of action is unknown. Here, by cryoEM of KSHV at 6-Å resolution, we show that SCP forms a crown on each hexon and uses a kinked helix to cross-link neighboring MCP subunits. SCP-null mutation decreased viral titer by 1,000 times and impaired but did not fully abolish capsid assembly, indicating an important but nonessential role of SCP. By truncating the C-terminal half of SCP and performing cryoEM reconstruction, we demonstrate that SCP's N-terminal half is responsible for the observed structure and function whereas the C-terminal half is flexible and dispensable. Serial truncations further highlight the critical importance of the N-terminal 10 aa, and cryoEM reconstruction of the one with six residues truncated localizes the N terminus of SCP in the cryoEM density map and enables us to construct a pseudoatomic model of SCP. Fitting of this SCP model and a homology model for the MCP upper domain into the cryoEM map reveals that SCP binds MCP largely via hydrophobic interactions and the kinked helix of SCP bridges over neighboring MCPs to form noncovalent cross-links. These data support a mechanistic model that tumor herpesvirus SCP reinforces the capsid for genome packaging, thus acting as a cementing protein similar to those found in many bacteriophages. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6213.map.gz emd_6213.map.gz | 387.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6213-v30.xml emd-6213-v30.xml emd-6213.xml emd-6213.xml | 9.6 KB 9.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6213.tif emd_6213.tif | 633.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6213 http://ftp.pdbj.org/pub/emdb/structures/EMD-6213 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6213 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6213 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6213.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6213.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM reconstruction of KSHV-SCPN86 mutant. The capsid structure is essentially identical to that of wild-type KSHV. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
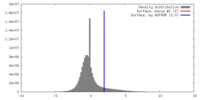
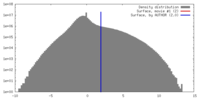
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Kaposi's sarcoma-associated herpesvirus (KSHV) mutant with the C-...
| Entire | Name: Kaposi's sarcoma-associated herpesvirus (KSHV) mutant with the C-terminal half of the smallest capsid protein (SCP) truncated |
|---|---|
| Components |
|
-Supramolecule #1000: Kaposi's sarcoma-associated herpesvirus (KSHV) mutant with the C-...
| Supramolecule | Name: Kaposi's sarcoma-associated herpesvirus (KSHV) mutant with the C-terminal half of the smallest capsid protein (SCP) truncated type: sample / ID: 1000 / Oligomeric state: icosahedral virus / Number unique components: 1 |
|---|
-Supramolecule #1: Human herpesvirus 8
| Supramolecule | Name: Human herpesvirus 8 / type: virus / ID: 1 / Name.synonym: Kaposi's sarcoma-associated herpesvirus Details: The smallest capsid protein (SCP) of the virus is truncated at residue number 86. NCBI-ID: 37296 / Sci species name: Human herpesvirus 8 / Sci species strain: BAC16 / Database: NCBI / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No / Syn species name: Kaposi's sarcoma-associated herpesvirus |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / synonym: VERTEBRATES Homo sapiens (human) / synonym: VERTEBRATES |
| Virus shell | Shell ID: 1 / Name: capsid / Diameter: 1350 Å / T number (triangulation number): 16 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: PBS |
|---|---|
| Grid | Details: Quantifoil R2/1-Cu200 |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | K2 camera super-resolution mode |
| Date | Nov 5, 2013 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Digitization - Sampling interval: 5.0 µm / Number real images: 1184 / Average electron dose: 25 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 48500 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 14000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each Particle |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 7.4 Å / Resolution method: OTHER / Software - Name: IMIRS, EMAN1 / Number images used: 1500 |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)