[English] 日本語
 Yorodumi
Yorodumi- EMDB-9312: Structure of the HO BMC shell: BMC-TD focused map, open inner por... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9312 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the HO BMC shell: BMC-TD focused map, open inner pore, compacted shell | |||||||||
 Map data Map data | Asymmetric reconstruction, BMC-T2, compacted shell | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | microcompartment / shell / compartmentalization / BMC fold / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Haliangium ochraceum (strain DSM 14365 / JCM 11303 / SMP-2) (bacteria) / Haliangium ochraceum (strain DSM 14365 / JCM 11303 / SMP-2) (bacteria) /  Haliangium ochraceum (bacteria) Haliangium ochraceum (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Greber BJ / Sutter M / Kerfeld CA | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2019 Journal: Structure / Year: 2019Title: The Plasticity of Molecular Interactions Governs Bacterial Microcompartment Shell Assembly. Authors: Basil J Greber / Markus Sutter / Cheryl A Kerfeld /  Abstract: Bacterial microcompartments (BMCs) are composed of an enzymatic core encapsulated by a selectively permeable protein shell that enhances catalytic efficiency. Many pathogenic bacteria derive ...Bacterial microcompartments (BMCs) are composed of an enzymatic core encapsulated by a selectively permeable protein shell that enhances catalytic efficiency. Many pathogenic bacteria derive competitive advantages from their BMC-based catabolism, implicating BMCs as drug targets. BMC shells are of interest for bioengineering due to their diverse and selective permeability properties and because they self-assemble. A complete understanding of shell composition and organization is a prerequisite for biotechnological applications. Here, we report the cryoelectron microscopy structure of a BMC shell at 3.0-Å resolution, using an image-processing strategy that allowed us to determine the previously uncharacterized structural details of the interactions formed by the BMC-T and BMC-T shell subunits in the context of the assembled shell. We found unexpected structural plasticity among these interactions, resulting in distinct shell populations assembled from varying numbers of the BMC-T and BMC-T subunits. We discuss the implications of these findings on shell assembly and function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9312.map.gz emd_9312.map.gz | 12.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9312-v30.xml emd-9312-v30.xml emd-9312.xml emd-9312.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9312.png emd_9312.png | 164.8 KB | ||
| Filedesc metadata |  emd-9312.cif.gz emd-9312.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9312 http://ftp.pdbj.org/pub/emdb/structures/EMD-9312 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9312 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9312 | HTTPS FTP |
-Related structure data
| Related structure data |  6n07MC  9296C  9307C  9308C  9309C  9310C  9311C  9313C  9314C  9315C  6mzuC  6mzvC  6mzxC  6mzyC  6n06C  6n09C  6n0fC  6n0gC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9312.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9312.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Asymmetric reconstruction, BMC-T2, compacted shell | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
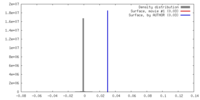
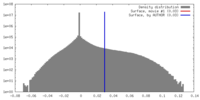
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bacterial microcompartment shell from Haliangium ochraceum
| Entire | Name: Bacterial microcompartment shell from Haliangium ochraceum |
|---|---|
| Components |
|
-Supramolecule #1: Bacterial microcompartment shell from Haliangium ochraceum
| Supramolecule | Name: Bacterial microcompartment shell from Haliangium ochraceum type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Haliangium ochraceum (strain DSM 14365 / JCM 11303 / SMP-2) (bacteria) Haliangium ochraceum (strain DSM 14365 / JCM 11303 / SMP-2) (bacteria) |
| Molecular weight | Theoretical: 6.5 MDa |
-Macromolecule #1: Microcompartments protein
| Macromolecule | Name: Microcompartments protein / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haliangium ochraceum (bacteria) / Strain: DSM 14365 / JCM 11303 / SMP-2 Haliangium ochraceum (bacteria) / Strain: DSM 14365 / JCM 11303 / SMP-2 |
| Molecular weight | Theoretical: 21.983164 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSITLRTYIF LDALQPQLAT FIGKTARGFL PVPGQASLWV EIAPGIAINR VTDAALKATK VQPAVQVVER AYGLLEVHHF DQGEVLAAG STILDKLEVR EEGRLKPQVM THQIIRAVEA YQTQIINRNS QGMMILPGES LFILETQPAG YAVLAANEAE K AANVHLVN ...String: MSITLRTYIF LDALQPQLAT FIGKTARGFL PVPGQASLWV EIAPGIAINR VTDAALKATK VQPAVQVVER AYGLLEVHHF DQGEVLAAG STILDKLEVR EEGRLKPQVM THQIIRAVEA YQTQIINRNS QGMMILPGES LFILETQPAG YAVLAANEAE K AANVHLVN VTPYGAFGRL YLAGSEAEID AAAEAAEAAI RSVSGVA UniProtKB: Bacterial microcompartment protein trimer-2 |
-Macromolecule #2: Microcompartments protein
| Macromolecule | Name: Microcompartments protein / type: protein_or_peptide / ID: 2 / Number of copies: 36 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haliangium ochraceum (strain DSM 14365 / JCM 11303 / SMP-2) (bacteria) Haliangium ochraceum (strain DSM 14365 / JCM 11303 / SMP-2) (bacteria)Strain: DSM 14365 / JCM 11303 / SMP-2 |
| Molecular weight | Theoretical: 10.126718 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MADALGMIEV RGFVGMVEAA DAMVKAAKVE LIGYEKTGGG YVTAVVRGDV AAVKAATEAG QRAAERVGEV VAVHVIPRPH VNVDAALPL GRTPGMDKSA UniProtKB: Bacterial microcompartment protein homohexamer |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Support film - #0 - Film type ID: 1 / Support film - #0 - Material: CARBON / Support film - #0 - topology: HOLEY / Support film - #1 - Film type ID: 2 / Support film - #1 - Material: CARBON / Support film - #1 - topology: CONTINUOUS / Details: unspecified | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: 5-7 second incubation of the sample on the grid before blotting and plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-30 / Number grids imaged: 1 / Number real images: 928 / Average exposure time: 4.5 sec. / Average electron dose: 25.0 e/Å2 Details: 928 images were retained after inspection for image quality. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 3.5 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 48543 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)