[English] 日本語
 Yorodumi
Yorodumi- EMDB-9255: Cryo-EM structure of ternary Csm-crRNA-target RNA with anti-tag s... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9255 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of ternary Csm-crRNA-target RNA with anti-tag sequence complex in type III-A CRISPR-Cas system | |||||||||
 Map data Map data | Ternary Csm-crRNA-target RNA with anti-tag sequence complex in type III-A CRISPR-Cas system | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cryo-EM structure / ternary Csm-crRNA-target RNA with anti-tag sequence complex / Type III CRISPR-Cas system / RNA BINDING PROTEIN-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationexonuclease activity / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / transferase activity / endonuclease activity / defense response to virus / Hydrolases; Acting on ester bonds / hydrolase activity / RNA binding / ATP binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |   Thermococcus onnurineus (archaea) Thermococcus onnurineus (archaea) | |||||||||
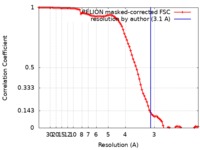
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Jia N / Wang C | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2019 Journal: Mol Cell / Year: 2019Title: Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity. Authors: Ning Jia / Charlie Y Mo / Chongyuan Wang / Edward T Eng / Luciano A Marraffini / Dinshaw J Patel /   Abstract: Type ΙΙΙ CRISPR-Cas systems provide robust immunity against foreign RNA and DNA by sequence-specific RNase and target RNA-activated sequence-nonspecific DNase and RNase activities. We report on ...Type ΙΙΙ CRISPR-Cas systems provide robust immunity against foreign RNA and DNA by sequence-specific RNase and target RNA-activated sequence-nonspecific DNase and RNase activities. We report on cryo-EM structures of Thermococcus onnurineus Csm binary, Csm-target RNA and Csm-target RNA ternary complexes in the 3.1 Å range. The topological features of the crRNA 5'-repeat tag explains the 5'-ruler mechanism for defining target cleavage sites, with accessibility of positions -2 to -5 within the 5'-repeat serving as sensors for avoidance of autoimmunity. The Csm3 thumb elements introduce periodic kinks in the crRNA-target RNA duplex, facilitating cleavage of the target RNA with 6-nt periodicity. Key Glu residues within a Csm1 loop segment of Csm adopt a proposed autoinhibitory conformation suggestive of DNase activity regulation. These structural findings, complemented by mutational studies of key intermolecular contacts, provide insights into Csm complex assembly, mechanisms underlying RNA targeting and site-specific periodic cleavage, regulation of DNase cleavage activity, and autoimmunity suppression. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9255.map.gz emd_9255.map.gz | 4.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9255-v30.xml emd-9255-v30.xml emd-9255.xml emd-9255.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9255_fsc.xml emd_9255_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_9255.png emd_9255.png | 95.4 KB | ||
| Filedesc metadata |  emd-9255.cif.gz emd-9255.cif.gz | 7.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9255 http://ftp.pdbj.org/pub/emdb/structures/EMD-9255 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9255 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9255 | HTTPS FTP |
-Related structure data
| Related structure data |  6mutMC  9253C  9254C  9256C  6muaC  6murC  6musC  6muuC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9255.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9255.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ternary Csm-crRNA-target RNA with anti-tag sequence complex in type III-A CRISPR-Cas system | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.089 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
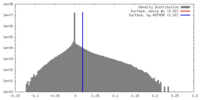
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : ternary Csm-crRNA-target RNA with anti-tag sequence complex
| Entire | Name: ternary Csm-crRNA-target RNA with anti-tag sequence complex |
|---|---|
| Components |
|
-Supramolecule #1: ternary Csm-crRNA-target RNA with anti-tag sequence complex
| Supramolecule | Name: ternary Csm-crRNA-target RNA with anti-tag sequence complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:   Thermococcus onnurineus (archaea) Thermococcus onnurineus (archaea) |
| Molecular weight | Theoretical: 250 KDa |
-Macromolecule #1: Uncharacterized protein Csm1
| Macromolecule | Name: Uncharacterized protein Csm1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermococcus onnurineus (archaea) Thermococcus onnurineus (archaea) |
| Molecular weight | Theoretical: 89.750602 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SQDPMEIDEL TALGGLLHDI GKPVQRAGLY SGDHSTQGAR FLRDLAENTG RAEYELLSLF SEFHHKGHMK NDELMIRRI KELSPERFGL TMEDVLNALW IVYEADNLAS GEREEGQPQA SRPLYSVFNP GKAYPWAELD FEKELPVPGD V FSIRSQDY ...String: MGSSHHHHHH SQDPMEIDEL TALGGLLHDI GKPVQRAGLY SGDHSTQGAR FLRDLAENTG RAEYELLSLF SEFHHKGHMK NDELMIRRI KELSPERFGL TMEDVLNALW IVYEADNLAS GEREEGQPQA SRPLYSVFNP GKAYPWAELD FEKELPVPGD V FSIRSQDY RELVKRLWEE LSKAKLRSDR LLPVLEKYLT FVSSVTSEGN IISLYDHMRM TSAIALAMLR AGCTAEDVRS GR CRKEKRF LLIEGDFSGI QDFIYRVSGK GTLKYLRARS AYLELIGWDV VLEILSRLGL TRANVVFNAG GHFMIIAQNT PDA VKELEE IRAKAVEWLY REFESDLYLA IEWEPVSGRE FGREGGKNLF AEARKRLKHK LTVRKLKRFG EIKGLFEHGH TERL AECPV CGRELPEGKL EPSASDPETK VCPTCNRLVS LGGNLPKLLG FGRTAKNDAG VLVEGPFSGF VPYLQGGRPV GEQIL VKNT LNPGEIPESA QFVPYFVADY FKKDPKGGVA TFEELSMAST GTRRLGVMKG DVDRLGEFFS SMDSPSKLAT ASRFMD YFF KGYIGAIIEG KFGYIIGDVP SLRDWPEEPD IVVVYAGGDD FFIVGAWDQI FELAFRVRRA FNAYTGGKLT LSVGLGY FD ERTPIYRMAD VVSERLDTAK DEGRNRVFVV GRSRPLDGKH KLSYEWNHYE ELWRTYAPRI YAGNGRLKGK LESKKGLL W KLLEIRELYV RDPNDVRWAY LTAYLLGRHG LSDLFPELVG IDTKAVERKE PQPVYWVDGV LKIVLMAVRR UniProtKB: CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A) |
-Macromolecule #2: Uncharacterized protein Csm2
| Macromolecule | Name: Uncharacterized protein Csm2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermococcus onnurineus (archaea) Thermococcus onnurineus (archaea) |
| Molecular weight | Theoretical: 21.210293 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SMAYHQKHGG YGRGGYGRQD RPQVDASRLF GESPDVVGIK KMLEGKGKQW EAIQPYFDNV VREAKNFLEW SPNKRLANAV TVAAYLTSQ GLKTNQVRKI LDMARTTELK VKRGEGDIKD DLVKMRYLLA YTVGKATGQS KYSLDAFHRI LDPMLEVLMG S PKKENFEK FYDFLQAVVA YHKFFGGGD UniProtKB: CRISPR system Cms protein Csm2 |
-Macromolecule #3: Uncharacterized protein Csm3
| Macromolecule | Name: Uncharacterized protein Csm3 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermococcus onnurineus (archaea) Thermococcus onnurineus (archaea) |
| Molecular weight | Theoretical: 32.765002 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SMDRRFYGKI VIKGKIKAVT GLHIGSQRDI SEIGGIANPV IKDPHTGLPY IPGSSLKGRL RSLFEILVNS RLGEWREKYP SLANYSPGS CRPDNQENCG KFFNRKINRG WIHVCPDYET ALACPVCRLF GASGKESNFP SRIIVRDAFL TKEWEEKWRA G EAITEAKI ...String: SMDRRFYGKI VIKGKIKAVT GLHIGSQRDI SEIGGIANPV IKDPHTGLPY IPGSSLKGRL RSLFEILVNS RLGEWREKYP SLANYSPGS CRPDNQENCG KFFNRKINRG WIHVCPDYET ALACPVCRLF GASGKESNFP SRIIVRDAFL TKEWEEKWRA G EAITEAKI EVGIDRVTSQ ANPRTNERVV AGAEFEFEII YNVENTTHWR DDIKNLLTAM ALLEDSYLGG SGSRGYGKVK FI FDSFEFR PLDYYRTGKD EDIVSIDARE KSVSDILSGF DSLFSEVEGK LEAG UniProtKB: CRISPR system Cms endoribonuclease Csm3 |
-Macromolecule #4: Uncharacterized protein Csm4
| Macromolecule | Name: Uncharacterized protein Csm4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermococcus onnurineus (archaea) Thermococcus onnurineus (archaea) |
| Molecular weight | Theoretical: 32.345061 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPKFIAVKLI PKGPFRDIPR ADTLFGAIGN AISAIHGQSA VEELVDAFVG GARISSAFPY SGDTYYLPKP LSVEPALEGI LTGLDEEER YTTAKRLRKA KYLDLKNFEL ALRLRPFTIP EEIPYARVDV PRVVLDRVTQ DSSIYFWEEI RFREKSGVYF L YSGPREVF ...String: MPKFIAVKLI PKGPFRDIPR ADTLFGAIGN AISAIHGQSA VEELVDAFVG GARISSAFPY SGDTYYLPKP LSVEPALEGI LTGLDEEER YTTAKRLRKA KYLDLKNFEL ALRLRPFTIP EEIPYARVDV PRVVLDRVTQ DSSIYFWEEI RFREKSGVYF L YSGPREVF DGYIAPAMRF LGDTGIGGKS TWGAGLFEVE FHEMKIDAPG SEYSVTLSNA LPTKTPVLWR LLRKGGWSFG RR KPRMTFI AEGSIVKNDP GGMERLELGL SHEVYVYGLT FPLGVELPEG LE UniProtKB: CRISPR system Cms protein Csm4 |
-Macromolecule #7: Uncharacterized protein Csm5
| Macromolecule | Name: Uncharacterized protein Csm5 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermococcus onnurineus (archaea) Thermococcus onnurineus (archaea) |
| Molecular weight | Theoretical: 46.091016 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTERTLKVLS PLHIGTGNEL TPVDIYPREN IIHVLDTERL VNDLMNLGVE LNEILALLKN PPGDAYIWKG YIEEFHLDPS DYSIYTLKI HGKIGRKSMQ IKEFIKLNGR PYIPGSSLKG AIRTAVLYKA LKECGDARAV MRVVSKVNGD VARDIGRSED V LDYYMSFL ...String: MTERTLKVLS PLHIGTGNEL TPVDIYPREN IIHVLDTERL VNDLMNLGVE LNEILALLKN PPGDAYIWKG YIEEFHLDPS DYSIYTLKI HGKIGRKSMQ IKEFIKLNGR PYIPGSSLKG AIRTAVLYKA LKECGDARAV MRVVSKVNGD VARDIGRSED V LDYYMSFL SRARIDRKRA DDLLEAIVFG MEPDRRSKIR YEPKRDPMKA LIVRDSKPVG RKHLAVYHVE VIGNPQPIPI WV EAIEPGA ATDVEIHVDT EALRLNADYF NGLLWECLKE RGEPGEVFED FLWEAVDEFY TAVMKYETIE VQKFGRYTSQ VRS FYASLE DHSGHVLRLG WGSGWLAMTI GLLLVEKGYK WENVRKKLGL GKKPGGSGFS REFPKTRRLA DGMPMGWVVL EHHH HHH UniProtKB: CRISPR system Cms protein Csm5 |
-Macromolecule #5: RNA (5'-R(*GP*UP*GP*GP*AP*AP*AP*GP*GP*CP*GP*GP*GP*CP*AP*GP*AP*GP*...
| Macromolecule | Name: RNA (5'-R(*GP*UP*GP*GP*AP*AP*AP*GP*GP*CP*GP*GP*GP*CP*AP*GP*AP*GP*GP*C)-3') type: rna / ID: 5 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Thermococcus onnurineus (archaea) Thermococcus onnurineus (archaea) |
| Molecular weight | Theoretical: 12.463438 KDa |
| Sequence | String: GUGGAAAGGC GGGCAGAGGC GGUUUGCGUA UUGGGCGC |
-Macromolecule #6: RNA (5'-R(P*CP*CP*UP*CP*UP*GP*CP*CP*CP*GP*CP*CP*UP*UP*UP*CP*CP*AP...
| Macromolecule | Name: RNA (5'-R(P*CP*CP*UP*CP*UP*GP*CP*CP*CP*GP*CP*CP*UP*UP*UP*CP*CP*AP*C)-3') type: rna / ID: 6 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Thermococcus onnurineus (archaea) Thermococcus onnurineus (archaea) |
| Molecular weight | Theoretical: 14.223495 KDa |
| Sequence | String: CCCUGGCGCC CAAUACGCAA ACCGCCUCUG CCCGCCUUUC CACGG |
-Macromolecule #8: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 8 / Number of copies: 3 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 8.8 / Component - Formula: Tris / Details: 20 mM Tris-HCl, pH 8.8, 250 mM NaCl, 2 mM DTT |
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.35 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)